| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,881,172 – 8,881,265 |
| Length | 93 |
| Max. P | 0.976004 |

| Location | 8,881,172 – 8,881,265 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 84.87 |
| Mean single sequence MFE | -29.00 |
| Consensus MFE | -23.70 |
| Energy contribution | -24.53 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.942179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8881172 93 + 22407834 GGGGCCGCCAUUAACGCAAAUGACUCUGCAAAAUGGCGGCCAUAUUAGGCAAGAAGACCAGGCCAAAACCCAAGAGCCA-------GUACCAGUAACACU ((((((((((((...(((........)))..))))))))))......(((..(.....)..)))...............-------...))......... ( -28.70) >DroSec_CAF1 91238 93 + 1 GGGGCCGCCAUUAACGCAAAUGACUCUGCAAAAUGGCGGCCAUAUUAGGCAAGAAGCCCAAGCCAAAACCCAAGAGCCA-------GUACCAGUAACACU ((((((((((((...(((........)))..))))))))))......(((.....))).....................-------...))......... ( -28.10) >DroSim_CAF1 89721 93 + 1 GGGGCCGCCAUUAACGCAAAUGACUCUGCAAAAUGGCGGCCAUAUUAGGCAAGAAGCCCAAGCCAAAACCCAAGAGCCA-------GUACCAGUAACACU ((((((((((((...(((........)))..))))))))))......(((.....))).....................-------...))......... ( -28.10) >DroEre_CAF1 95717 99 + 1 GGGGCCGCCAUUAACGCAAAUGACUCUGCAAAAUGGCGGCCGUAUUAGGCAAGAAGCCCAGGCCAAAACCCAAGAGCCAGCA-CCAGUACCAGUAAAACU ((((((((((((...(((........)))..))))))))))((....(((.....)))..((((.........).))).)).-))............... ( -31.70) >DroYak_CAF1 94247 100 + 1 GGGGCCGCCAUUAACGCAAAUGACUCUGCAAAAUGGCGGCCAUAUUAGGCAAGAAGCCCAGGCCAAAGCCCAAGAGCCAGUAUCCAGUACCAGUAACACU ((((((((((((...(((........)))..))))))))))(((((.(((..(.....).(((....))).....))))))))......))......... ( -32.00) >DroPer_CAF1 123579 77 + 1 GGAGCCGCCAUUAACAGAGAUGACUCUGCAAAAUGGCGGCCAUAUUAGGCAAGAGAAGCAAG--AGGAGCCAAGAGUCA--------------------- ((..((((((((..(((((....)))))...)))))).(((......)))............--.))..))........--------------------- ( -25.40) >consensus GGGGCCGCCAUUAACGCAAAUGACUCUGCAAAAUGGCGGCCAUAUUAGGCAAGAAGCCCAAGCCAAAACCCAAGAGCCA_______GUACCAGUAACACU ((((((((((((...(((........)))..))))))))))......(((..(.....)..))).....))............................. (-23.70 = -24.53 + 0.83)

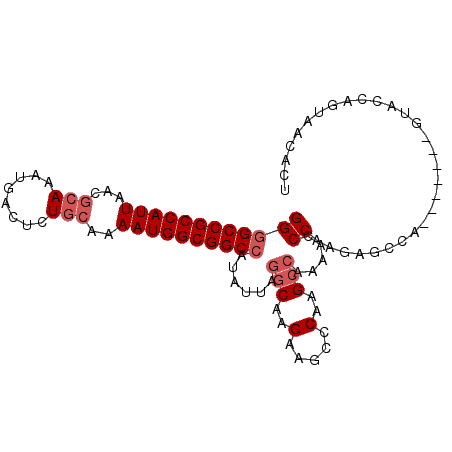
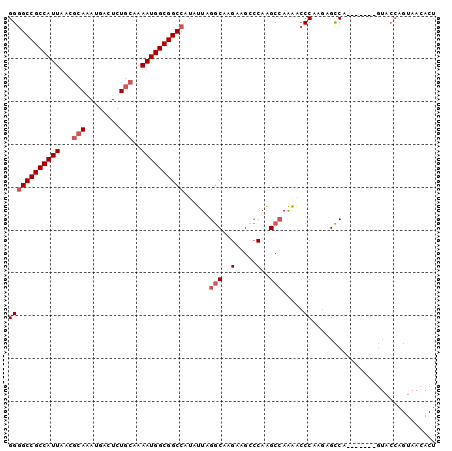
| Location | 8,881,172 – 8,881,265 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 84.87 |
| Mean single sequence MFE | -37.95 |
| Consensus MFE | -30.50 |
| Energy contribution | -30.95 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.16 |
| Mean z-score | -4.18 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8881172 93 - 22407834 AGUGUUACUGGUAC-------UGGCUCUUGGGUUUUGGCCUGGUCUUCUUGCCUAAUAUGGCCGCCAUUUUGCAGAGUCAUUUGCGUUAAUGGCGGCCCC .((((((..((((.-------.((((...((((....))))))))....))))))))))((((((((((.((((((....))))))..)))))))))).. ( -37.40) >DroSec_CAF1 91238 93 - 1 AGUGUUACUGGUAC-------UGGCUCUUGGGUUUUGGCUUGGGCUUCUUGCCUAAUAUGGCCGCCAUUUUGCAGAGUCAUUUGCGUUAAUGGCGGCCCC .((((((..((((.-------.(((((...(((....))).)))))...))))))))))((((((((((.((((((....))))))..)))))))))).. ( -38.60) >DroSim_CAF1 89721 93 - 1 AGUGUUACUGGUAC-------UGGCUCUUGGGUUUUGGCUUGGGCUUCUUGCCUAAUAUGGCCGCCAUUUUGCAGAGUCAUUUGCGUUAAUGGCGGCCCC .((((((..((((.-------.(((((...(((....))).)))))...))))))))))((((((((((.((((((....))))))..)))))))))).. ( -38.60) >DroEre_CAF1 95717 99 - 1 AGUUUUACUGGUACUGG-UGCUGGCUCUUGGGUUUUGGCCUGGGCUUCUUGCCUAAUACGGCCGCCAUUUUGCAGAGUCAUUUGCGUUAAUGGCGGCCCC ..........(((.(((-.((.(((((..((((....)))))))))....))))).)))((((((((((.((((((....))))))..)))))))))).. ( -41.50) >DroYak_CAF1 94247 100 - 1 AGUGUUACUGGUACUGGAUACUGGCUCUUGGGCUUUGGCCUGGGCUUCUUGCCUAAUAUGGCCGCCAUUUUGCAGAGUCAUUUGCGUUAAUGGCGGCCCC .((((((..((((..(((....(((((..((((....))))))))))))))))))))))((((((((((.((((((....))))))..)))))))))).. ( -42.50) >DroPer_CAF1 123579 77 - 1 ---------------------UGACUCUUGGCUCCU--CUUGCUUCUCUUGCCUAAUAUGGCCGCCAUUUUGCAGAGUCAUCUCUGUUAAUGGCGGCUCC ---------------------........(((....--............)))......((((((((((..((((((....)))))).)))))))))).. ( -29.09) >consensus AGUGUUACUGGUAC_______UGGCUCUUGGGUUUUGGCCUGGGCUUCUUGCCUAAUAUGGCCGCCAUUUUGCAGAGUCAUUUGCGUUAAUGGCGGCCCC ...........................((((((...(((....)))....))))))...((((((((((.((((((....))))))..)))))))))).. (-30.50 = -30.95 + 0.45)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:50 2006