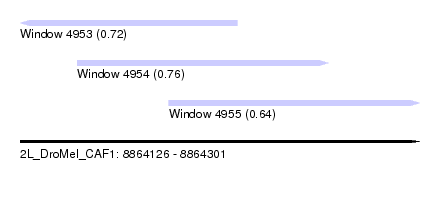
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,864,126 – 8,864,301 |
| Length | 175 |
| Max. P | 0.755266 |

| Location | 8,864,126 – 8,864,221 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 70.60 |
| Mean single sequence MFE | -28.78 |
| Consensus MFE | -14.57 |
| Energy contribution | -14.05 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.723949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
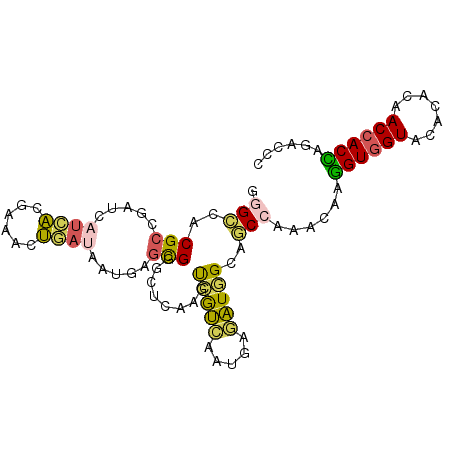
>2L_DroMel_CAF1 8864126 95 - 22407834 GGGCCACGCCGAUCAUCACGAAACUGAUAAUGAGCGGGCUCAAGUCGUGGAUGAAAUGGCAGCCAAACAAGGUGGUACACACAACCACCAGACCU (((((.(((..((.((((......)))).))..))))))))..(((.(((.((......)).))).....((((((.......)))))).))).. ( -31.10) >DroSec_CAF1 74363 95 - 1 GGGCCACGCCGAUCAUCACGAAACUAAUAGUGAGCGGGCUCAAAUCGUCAAUGAGAUGGCAGCCAAACAAGGUGGUACACACAACCACCAGACCC (((((.(((......((((..........)))))))))))).....(((.......(((...))).....((((((.......)))))).))).. ( -27.70) >DroSim_CAF1 72867 95 - 1 AGGCCACGCCGAUCAUCACGAAACUGAUAGUGAGCGGGCUCAAAUCGUCAAUGAGAUGGCAGCCAAACAAGGUGGUACACACAACCACCAGACCC .(((...((((.((((.((((.((.....))(((....)))...))))..))))..)))).)))......((((((.......))))))...... ( -27.80) >DroEre_CAF1 78630 95 - 1 GGGCCACGCCGAUCAUCACGAAGCUGACGACGAGUGUGCUCAACUUGUCGAUAAGAUAGCAACCGAACAAAGUGGUGCAGACAACCACUACAGCC .(((...)))............((((.((((((((.......))))))))....................((((((.......)))))).)))). ( -27.30) >DroYak_CAF1 76576 95 - 1 GCGCCACGCCGAUCAUCACGCCAAUGAUGACGAACGUGGUCAAGUCGUCAAUCAGAUAGGUGCCGAACAAGGUGGUGCACACAACCACCACGCUA ((((((((.((.((((((......))))))))..))))))....(((...(((.....)))..)))....((((((.......))))))..)).. ( -30.00) >DroAna_CAF1 76046 95 - 1 GAGUCACGCCGAAGACGGCCAGAACCAAGGGCAGCGUCCCUAGGCCAGUAGCGGAUUGGGCACCACCCAACGUAGUCAUGGCCACCACGAAGCAU (..((..((((....))))........((((......)))).(((((...(((...((((.....)))).))).....))))).....))..).. ( -28.80) >consensus GGGCCACGCCGAUCAUCACGAAACUGAUAAUGAGCGGGCUCAAGUCGUCAAUGAGAUGGCAGCCAAACAAGGUGGUACACACAACCACCAGACCC .(((..(((.....((((......)))).....)))........(((((.....)))))..)))......((((((.......))))))...... (-14.57 = -14.05 + -0.52)

| Location | 8,864,151 – 8,864,261 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 74.67 |
| Mean single sequence MFE | -34.15 |
| Consensus MFE | -22.47 |
| Energy contribution | -21.09 |
| Covariance contribution | -1.38 |
| Combinations/Pair | 1.63 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8864151 110 + 22407834 UUGUUUGGCUGCCAUUUCAUCCACGACUUGAGCCCGCUCAUUAUCAGUUUCGUGAUGAUCGGCGUGGCCCUGAUCAUUAUGUUACUCAACUUCUCUGGGGCCAUGUGCCC ......(((.(((...(((((.((((((((((....)))).....))..)))))))))..)))((((((((........((.....))........))))))))..))). ( -33.69) >DroSec_CAF1 74388 110 + 1 UUGUUUGGCUGCCAUCUCAUUGACGAUUUGAGCCCGCUCACUAUUAGUUUCGUGAUGAUCGGCGUGGCCCUGAUCAUUCUGUUUCUCAACUUCUCAGGGGCCAUGUGCCC ......(((.((((((((((.((((((.((((....))))..))).)))..)))).))).)))((((((((........((.....))........))))))))..))). ( -33.49) >DroSim_CAF1 72892 110 + 1 UUGUUUGGCUGCCAUCUCAUUGACGAUUUGAGCCCGCUCACUAUCAGUUUCGUGAUGAUCGGCGUGGCCUUGAUCAUUCUGUUUCUCAACUUCUCAGGGGCCAUGUGCCC ......(((.((((((((((.((((((.((((....))))..))).)))..)))).))).)))((((((((........((.....))........))))))))..))). ( -32.79) >DroEre_CAF1 78655 110 + 1 UUGUUCGGUUGCUAUCUUAUCGACAAGUUGAGCACACUCGUCGUCAGCUUCGUGAUGAUCGGCGUGGCCCUGAUCAUCGUGAUUCUUAACUUUUCGGGGGCUAUGUGCCC ...................((((.((((((((..(((.((..(....)..)).(((((((((.(....)))))))))))))...)))))))).))))((((.....)))) ( -31.90) >DroYak_CAF1 76601 110 + 1 UUGUUCGGCACCUAUCUGAUUGACGACUUGACCACGUUCGUCAUCAUUGGCGUGAUGAUCGGCGUGGCGCUGAUCAUACUGAUUCUCAACUUCGCGGGUGCCAUGUGUCC ......(((((((...((((.((((((........).)))))))))...(((..((((((((((...))))))))))..((.....))....))))))))))........ ( -37.90) >DroAna_CAF1 76071 110 + 1 UUGGGUGGUGCCCAAUCCGCUACUGGCCUAGGGACGCUGCCCUUGGUUCUGGCCGUCUUCGGCGUGACUCUCAUCGUCCUCAUUCUUAACUUUUCUGGAGCGAUGUGCCC ..(((..((((((...(((....)))....))).)))..)))..(((....((((....))))........((((((((.................))).))))).))). ( -35.13) >consensus UUGUUUGGCUGCCAUCUCAUUGACGACUUGAGCACGCUCACCAUCAGUUUCGUGAUGAUCGGCGUGGCCCUGAUCAUUCUGAUUCUCAACUUCUCAGGGGCCAUGUGCCC ......(((..............(((..((((....)))).(((((......))))).)))((((((((((........((.....))........))))))))))))). (-22.47 = -21.09 + -1.38)


| Location | 8,864,191 – 8,864,301 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.38 |
| Mean single sequence MFE | -40.63 |
| Consensus MFE | -26.60 |
| Energy contribution | -26.55 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
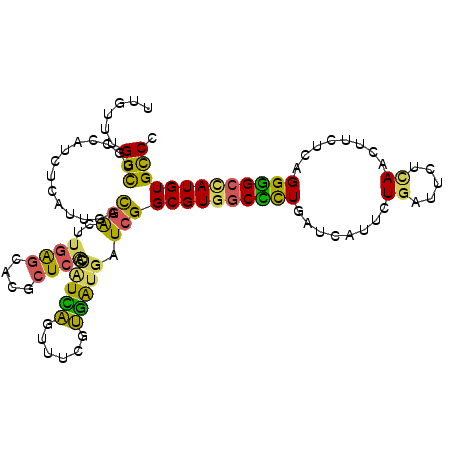
>2L_DroMel_CAF1 8864191 110 + 22407834 UUAUC-AGUUUCGUGAUGAUCGGCGUGGCCC---UGAUCAUUAUGUUACUCAACUUCUCUGGGGCCAUGUGCCCGCAGGAGUUCCUGCCCGGCGGUGUUUGCUCCACGCUGCUG .....-(((..((((((((((((.(....))---)))))))))))..)))...........((((.....))))(((((....))))).(((((((((.......))))))))) ( -42.90) >DroSec_CAF1 74428 110 + 1 CUAUU-AGUUUCGUGAUGAUCGGCGUGGCCC---UGAUCAUUCUGUUUCUCAACUUCUCAGGGGCCAUGUGCCCGCAGGAGUUCCUGCCCGGCGGUGUUUGCUCCACGCUGCUG ....(-(((..((((.......(((((((((---(........((.....))........))))))))))(((.(((((....)))))..)))...........))))..)))) ( -39.09) >DroSim_CAF1 72932 110 + 1 CUAUC-AGUUUCGUGAUGAUCGGCGUGGCCU---UGAUCAUUCUGUUUCUCAACUUCUCAGGGGCCAUGUGCCCGCAGGAGUUCCUGCCCGGCGGUGUUUGCUCCACGCUGCUG ....(-(((..((((.......(((((((((---(........((.....))........))))))))))(((.(((((....)))))..)))...........))))..)))) ( -37.79) >DroEre_CAF1 78695 110 + 1 UCGUC-AGCUUCGUGAUGAUCGGCGUGGCCC---UGAUCAUCGUGAUUCUUAACUUUUCGGGGGCUAUGUGCCCACAGGAGUUCCUGCCCGGCGGUGUCUGUUCCACGCUGCUA (((((-(......)))))).((((((((..(---.((.((((((..............(((((((.....)))).((((....)))).))))))))))).)..))))))))... ( -42.23) >DroWil_CAF1 124285 110 + 1 CUGUGGUGCUGA----UGGUGGUGCUGGCUACAGUCGUGGUGUUGAUAUUGAAUUUCUCCGGGGCCAAGUGCCCGCAGCAGCUUCUGCCGGGCGGAAUCUGCUCCACUUCUCUG ..((((.((.((----(..((((.((((((((....)))))...((.........))..))).))))..((((((..((((...))))))))))..))).)).))))....... ( -37.40) >DroYak_CAF1 76641 110 + 1 UCAUC-AUUGGCGUGAUGAUCGGCGUGGCGC---UGAUCAUACUGAUUCUCAACUUCGCGGGUGCCAUGUGUCCGCAGGAGUUCCUGCCCGGUGGUGUCUGUUCCACGCUGCUA (((((-((....))))))).((((((((.((---.(((..(((((......(((((((((((..(...)..))))).))))))......)))))..))).)).))))))))... ( -44.40) >consensus CUAUC_AGUUUCGUGAUGAUCGGCGUGGCCC___UGAUCAUUCUGAUUCUCAACUUCUCAGGGGCCAUGUGCCCGCAGGAGUUCCUGCCCGGCGGUGUCUGCUCCACGCUGCUG ......................(((((((((....((..................))....)))))))))....(((((....)))))..((((((((.......)))))))). (-26.60 = -26.55 + -0.05)

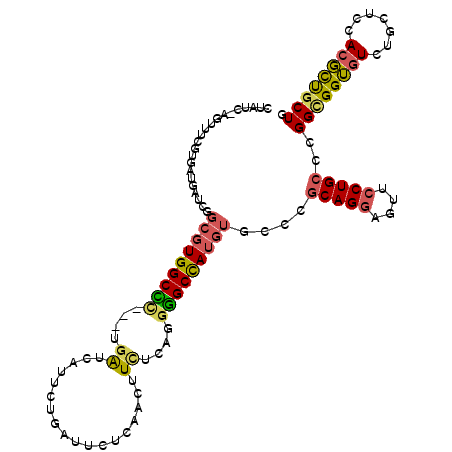
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:42 2006