| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
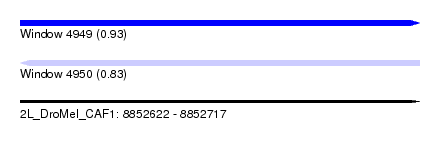
| Location | 8,852,622 – 8,852,717 |
| Length | 95 |
| Max. P | 0.931367 |

| Location | 8,852,622 – 8,852,717 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 80.26 |
| Mean single sequence MFE | -30.93 |
| Consensus MFE | -18.78 |
| Energy contribution | -19.28 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
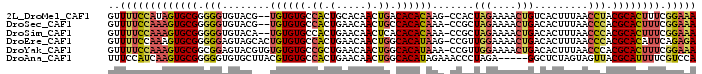
>2L_DroMel_CAF1 8852622 95 + 22407834 GUUUUCCAUAGUGCGGGGGUGUACG--UGUGUGCCACUGCACAACUGACACACAAG-CCACUAGAAAACUGUCACUUUAACCUACGCACUUUCGGAAA ..(((((..((((((..(((....(--(.(((((....)))))))(((((......-............))))).....)))..))))))...))))) ( -27.87) >DroSec_CAF1 62796 95 + 1 GUUUUCCAAAGUGCGGGGGUGUACG--UGUGUGCCACUGAACAACUGCCACACAAA-CCGCUAGAAAACUGACACUUUAACCCACGCACUUUCGGAAA ..(((((((((((((((((((....--((((((.((.(.....).)).))))))..-.)))(((....))).........))).)))))))).))))) ( -31.10) >DroSim_CAF1 61360 95 + 1 GUUUUCCAAAGUGCGGGGGUGUACA--UGUGUGCCACUGAACAACUCACACACAAA-CCGCUAGAAAACUGACACUUUAACCCACGCACUUUCGGAAA ..(((((((((((((.((((.....--((((((....(((.....)))))))))..-..(.(((....))).)......)))).)))))))).))))) ( -29.90) >DroEre_CAF1 65662 97 + 1 GUUUUCCAAAGUGCGGGGGAGUAGCACUGUGUGCCACUGAACAACUGGCACAUAAG-CCGUUGGAAAACUGACACUUUAACCCACGCACAUUCAGAGA ..........((((((((((((.((..(((((((((.(.....).))))))))).)-).((..(....)..))))))...))).)))))......... ( -32.20) >DroYak_CAF1 64643 97 + 1 GUUUUCCAAAGUGCGGCGGAGUACGUGUGUGUGCCGCUGAACAACUGGCACAUAAA-CCGUUGGAAAACUGACACUUUAACCCACGCACUUUCGGAAA ..(((((((((((((..((.((..((.(((((((((.(.....).))))))))).)-).((..(....)..))......)))).)))))))).))))) ( -35.60) >DroAna_CAF1 65324 93 + 1 UUUCCAUCAAGUGCGGGGGUGUGCUUACGUGUGCCACUGAACAACUGGCACAUAGAAACCCUAGA-----GGCUCUAGUAGUUACGCAUUUUCGUCCA ..........(..((..((((((...((((((((((.(.....).)))))))).......((((.-----....))))..))..))))))..))..). ( -28.90) >consensus GUUUUCCAAAGUGCGGGGGUGUACG__UGUGUGCCACUGAACAACUGGCACACAAA_CCGCUAGAAAACUGACACUUUAACCCACGCACUUUCGGAAA ..(((((((((((((.(((........((((((.((.(.....).)).)))))).......(((....))).........))).)))))))).))))) (-18.78 = -19.28 + 0.50)
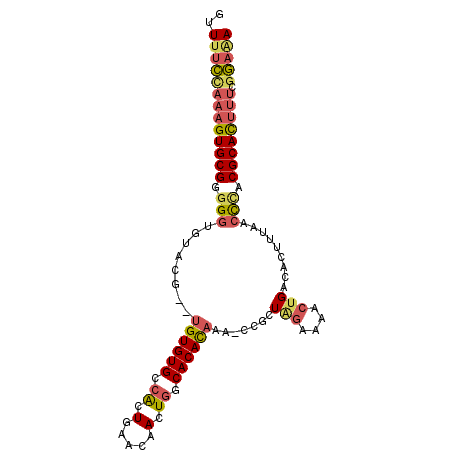
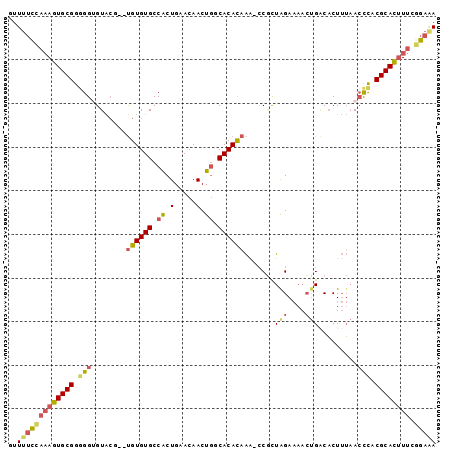
| Location | 8,852,622 – 8,852,717 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 80.26 |
| Mean single sequence MFE | -32.78 |
| Consensus MFE | -17.91 |
| Energy contribution | -18.91 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8852622 95 - 22407834 UUUCCGAAAGUGCGUAGGUUAAAGUGACAGUUUUCUAGUGG-CUUGUGUGUCAGUUGUGCAGUGGCACACA--CGUACACCCCCGCACUAUGGAAAAC ((((((..((((((..(((..((((.((((....)).)).)-)))((((((((.(.....).)))))))).--.....)))..)))))).)))))).. ( -34.50) >DroSec_CAF1 62796 95 - 1 UUUCCGAAAGUGCGUGGGUUAAAGUGUCAGUUUUCUAGCGG-UUUGUGUGGCAGUUGUUCAGUGGCACACA--CGUACACCCCCGCACUUUGGAAAAC (((((.((((((((.((((.(((.((..((....))..)).-)))(((((.((.(.....).)).))))).--.....)))).))))))))))))).. ( -36.10) >DroSim_CAF1 61360 95 - 1 UUUCCGAAAGUGCGUGGGUUAAAGUGUCAGUUUUCUAGCGG-UUUGUGUGUGAGUUGUUCAGUGGCACACA--UGUACACCCCCGCACUUUGGAAAAC (((((.((((((((.((((.(((.((..((....))..)).-)))((((((.(.(.....).).)))))).--.....)))).))))))))))))).. ( -34.80) >DroEre_CAF1 65662 97 - 1 UCUCUGAAUGUGCGUGGGUUAAAGUGUCAGUUUUCCAACGG-CUUAUGUGCCAGUUGUUCAGUGGCACACAGUGCUACUCCCCCGCACUUUGGAAAAC ..((..((.(((((.(((.................((((((-(......))).))))...((((((((...)))))))).))))))))))..)).... ( -35.30) >DroYak_CAF1 64643 97 - 1 UUUCCGAAAGUGCGUGGGUUAAAGUGUCAGUUUUCCAACGG-UUUAUGUGCCAGUUGUUCAGCGGCACACACACGUACUCCGCCGCACUUUGGAAAAC (((((.(((((((((((((....((((..(((....)))..-....((((((.(((....))))))))).))))..)).))).))))))))))))).. ( -35.30) >DroAna_CAF1 65324 93 - 1 UGGACGAAAAUGCGUAACUACUAGAGCC-----UCUAGGGUUUCUAUGUGCCAGUUGUUCAGUGGCACACGUAAGCACACCCCCGCACUUGAUGGAAA ..........((((......((((....-----.)))).((((.(.(((((((.(.....).))))))).).)))).......))))........... ( -20.70) >consensus UUUCCGAAAGUGCGUGGGUUAAAGUGUCAGUUUUCUAGCGG_UUUAUGUGCCAGUUGUUCAGUGGCACACA__CGUACACCCCCGCACUUUGGAAAAC ((((((((.(((((((((...............)))).........(((((((.(.....).)))))))..............))))))))))))).. (-17.91 = -18.91 + 1.00)

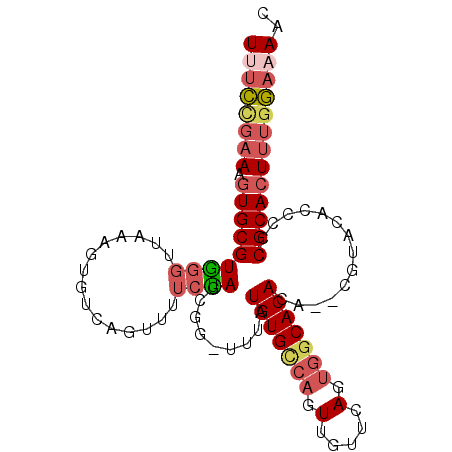
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:38 2006