
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,842,128 – 8,842,234 |
| Length | 106 |
| Max. P | 0.538234 |

| Location | 8,842,128 – 8,842,234 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.23 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -15.43 |
| Energy contribution | -15.85 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8842128 106 + 22407834 GGCA---GCAGCGAAAAGCAACAACAACACUGGGAGCACAAAAGGUGUGCACCAUAAUGGCUGACAAAAAGUCGGCCUUAGCCUUUAAUAGCA-CUGAGUUUAACAGCCA (((.---...((.(((.((...........(((..(((((.....))))).)))....(((((((.....)))))))...)).)))....)).-....(.....).))). ( -30.00) >DroPse_CAF1 72310 104 + 1 ---A---ACAACAACAUUCAGCAGCAGCAGCAGAAGCACAAAAGGUGUGCACCAUAAUGGCUGACAAAAAGUCGGCCUUAGCCUUUAAUAGCCCCAGAGUUUAACAGCCA ---.---........((((.((....)).((....(((((.....)))))........(((((((.....)))))))...))..............)))).......... ( -22.80) >DroSec_CAF1 52345 106 + 1 GGCA---GCAGCGAACAGCAACAAUAACACUGGGAGCACAAAAGGUAUGCACCAUAAUGGCUGACAAAAAGUCGGCCUUAGCCUUUAAUAGCA-CUGAGUUUAACAGCCA (((.---...((.....)).........(((.((.((...(((((((((...)))...(((((((.....)))))))...))))))....)).-)).)))......))). ( -29.30) >DroYak_CAF1 53768 108 + 1 GGCAGAAGCAGCA-AAAGCAACAACAACACUGGGAGCACAAAAGGUGCGCACCAUAAUGGCUGACAAAAAGUCGGCCUUAGCCUUUAAUAGCA-CUGAGUUUAACUGCCA (((((.....((.-...)).........(((.((.((......(((....))).....(((((((.....))))))).............)).-)).)))....))))). ( -32.70) >DroAna_CAF1 54593 96 + 1 ---A---GC-------AGCAACAACAACACUGGGAGCACAAAAGGUGUGCACUAUAAUGGCUGACAAAAAGUCGGCCUUAGCCUUUAAUAGCA-CAGAGUUUAACAGCCA ---.---..-------.((......(((.(((((.(((((.....))))).)).....(((((((.....)))))))...((........)).-))).))).....)).. ( -23.70) >DroPer_CAF1 74186 104 + 1 ---A---ACGACAACAUUCGUCAACAGCCCGAGAUGCUCACAAGGUGUGCACGAUACUGCUCGACAGAAAAUCGGCCUUAGCCUUUAAUAGCCCCAGAGAUGAACAGCCA ---.---.........((((((........(((...)))..(((((..((.((((.(((.....)))...))))))....))))).............))))))...... ( -18.10) >consensus ___A___GCAGCAAAAAGCAACAACAACACUGGGAGCACAAAAGGUGUGCACCAUAAUGGCUGACAAAAAGUCGGCCUUAGCCUUUAAUAGCA_CAGAGUUUAACAGCCA ...................(((..........((.(((((.....))))).)).....(((((((.....))))))).....................)))......... (-15.43 = -15.85 + 0.42)

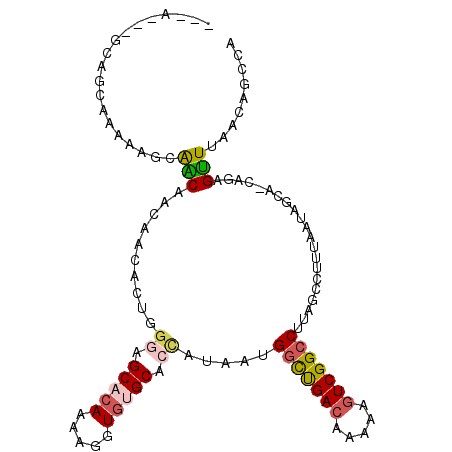
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:33 2006