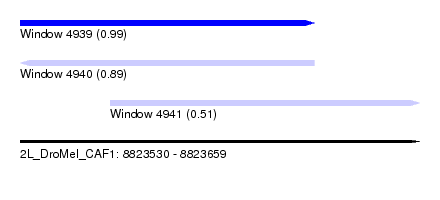
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,823,530 – 8,823,659 |
| Length | 129 |
| Max. P | 0.992906 |

| Location | 8,823,530 – 8,823,625 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.29 |
| Mean single sequence MFE | -27.38 |
| Consensus MFE | -15.97 |
| Energy contribution | -16.93 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.992906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
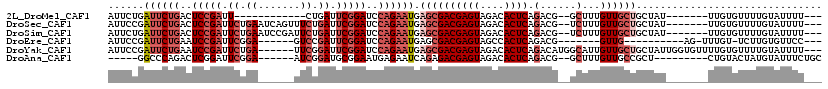
>2L_DroMel_CAF1 8823530 95 + 22407834 ---AAAAUACAAAACACAA-------AUAGCAGCAACAAAGC--CGUCUGAGUGUCUACUCGUCGCUCAUUCUGGAUCCGAAUCAG------------AAUCGGAGUCAGAAUCAGAAU ---................-------...((.((......))--.)).((((((.(.....).))))))(((((..(((((.....------------..)))))..)))))....... ( -21.60) >DroSec_CAF1 33316 107 + 1 ---AAAAUACAAAACACAA-------AUAGCAGCAACAAAGA--CGUCUGAGUGUCUACUCGUCGCUCAUUCUGGAUCCGAAUCAGAAACUGAUUCAGAAUCGGAGUCAGAAUCGGAAU ---................-------..............((--(...((((((.(.....).)))))).(((((.((.(((((((...))))))).)).))))))))........... ( -24.50) >DroSim_CAF1 31523 107 + 1 ---AAAAUACAAAACACAA-------AUAGCAGCAACAAAGA--CGUCUGAGUGUCUACUCGUCGCUCAUUCUGGAUCCGAAUCAGAAUCGGAUUCAGAAUCGGAGUCAGAAUCAGAAU ---................-------................--..(((((...(((((((..((...(((((((((((((.......))))))))))))))))))).))).))))).. ( -29.60) >DroEre_CAF1 36187 91 + 1 ---GGAACACAAGA-ACAAA-CU----------CAAC-------CGUCUGAGUGGCUACUCGUCGCUCAUUCUGGAUCCGAAUCGGAC------UCCGAAUCGGAUUCAGAAUCGGAAU ---...........-.....-..----------...(-------((...(((((((.....)))))))(((((((((((((.((((..------.)))).))))))))))))))))... ( -36.90) >DroYak_CAF1 34419 110 + 1 ---AAAAUACAAAACACAAAACACCAAUAGCAGCAACAAUGCCAUGUCUGAGUGUCUACUCGUCGCUCAUUCUGGAUCCGAAUCCGAA------UCAGAAUCGGAUUCAGAAUCGGAAU ---....................((....(((.......)))......((((((.(.....).))))))((((((((((((.((....------...)).))))))))))))..))... ( -28.90) >DroAna_CAF1 33641 97 + 1 GCAGAAAUACAUAGUACAG---------AGCGGCAACAAAGC--CGUCUGAGUGUCUACUCGUCUCUGAUUCUCAUUCCGCAUCCGAU------UCCGAAUCCGAGUCUGGGCC----- ((.(((......(((.(((---------((((((......))--))...((((....))))..))))))))....))).)).((((((------((.......))))).)))..----- ( -22.80) >consensus ___AAAAUACAAAACACAA_______AUAGCAGCAACAAAGC__CGUCUGAGUGUCUACUCGUCGCUCAUUCUGGAUCCGAAUCAGAA______UCAGAAUCGGAGUCAGAAUCGGAAU ................................................((((((.(.....).))))))((((((.(((((.((.((.......)).)).))))).))))))....... (-15.97 = -16.93 + 0.96)


| Location | 8,823,530 – 8,823,625 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.29 |
| Mean single sequence MFE | -30.33 |
| Consensus MFE | -15.55 |
| Energy contribution | -17.62 |
| Covariance contribution | 2.07 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8823530 95 - 22407834 AUUCUGAUUCUGACUCCGAUU------------CUGAUUCGGAUCCAGAAUGAGCGACGAGUAGACACUCAGACG--GCUUUGUUGCUGCUAU-------UUGUGUUUUGUAUUUU--- ...((.((((((..(((((..------------.....)))))..)))))).))..((((...(((((..((.((--((......))))))..-------..))))))))).....--- ( -27.10) >DroSec_CAF1 33316 107 - 1 AUUCCGAUUCUGACUCCGAUUCUGAAUCAGUUUCUGAUUCGGAUCCAGAAUGAGCGACGAGUAGACACUCAGACG--UCUUUGUUGCUGCUAU-------UUGUGUUUUGUAUUUU--- ......((((((..(((((.((.(((.....))).)).)))))..)))))).((((((((..((((........)--))))))))))).....-------................--- ( -30.30) >DroSim_CAF1 31523 107 - 1 AUUCUGAUUCUGACUCCGAUUCUGAAUCCGAUUCUGAUUCGGAUCCAGAAUGAGCGACGAGUAGACACUCAGACG--UCUUUGUUGCUGCUAU-------UUGUGUUUUGUAUUUU--- ......((((((..(((((.((.((((...)))).)).)))))..)))))).((((((((..((((........)--))))))))))).....-------................--- ( -29.70) >DroEre_CAF1 36187 91 - 1 AUUCCGAUUCUGAAUCCGAUUCGGA------GUCCGAUUCGGAUCCAGAAUGAGCGACGAGUAGCCACUCAGACG-------GUUG----------AG-UUUGU-UCUUGUGUUCC--- ......((((((.((((((.((((.------..)))).)))))).))))))((((.(((((.(((.((((((...-------.)))----------))-)..))-)))))))))).--- ( -35.30) >DroYak_CAF1 34419 110 - 1 AUUCCGAUUCUGAAUCCGAUUCUGA------UUCGGAUUCGGAUCCAGAAUGAGCGACGAGUAGACACUCAGACAUGGCAUUGUUGCUGCUAUUGGUGUUUUGUGUUUUGUAUUUU--- .....(..(((((((((((......------.)))))))))))..)((((((...((((...((((((.(((...(((((....)))))...)))))))))..))))...))))))--- ( -32.70) >DroAna_CAF1 33641 97 - 1 -----GGCCCAGACUCGGAUUCGGA------AUCGGAUGCGGAAUGAGAAUCAGAGACGAGUAGACACUCAGACG--GCUUUGUUGCCGCU---------CUGUACUAUGUAUUUCUGC -----....((...(((.(((((..------..))))).)))..)).....((((((...((((.....((((((--((......)))).)---------)))..))))...)))))). ( -26.90) >consensus AUUCCGAUUCUGACUCCGAUUCUGA______UUCUGAUUCGGAUCCAGAAUGAGCGACGAGUAGACACUCAGACG__GCUUUGUUGCUGCUAU_______UUGUGUUUUGUAUUUU___ ......((((((..(((((.((.((.......)).)).)))))..)))))).((((((((((....)))).(......)...))))))............................... (-15.55 = -17.62 + 2.07)
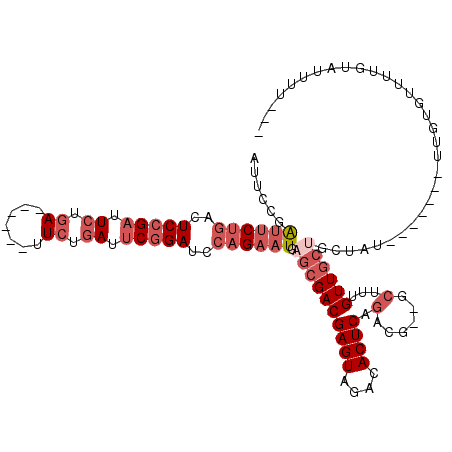
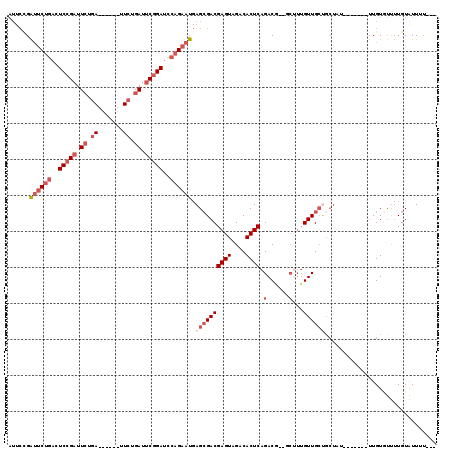
| Location | 8,823,559 – 8,823,659 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.62 |
| Mean single sequence MFE | -34.25 |
| Consensus MFE | -15.35 |
| Energy contribution | -16.78 |
| Covariance contribution | 1.43 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.45 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8823559 100 + 22407834 AGC--CGUCUGAGUGUCUACUCGUCGCUCAUUCUGGAUCCGAAUCAG------------AAUCGGAGUCAGAAUCAGAAUCGCAAUCCGAGUAUGAGUCUG------AGUAUAAGUCUGG ..(--((..((((((.(.....).))))))..).))........(((------------(.....(.(((((.(((...(((.....)))...))).))))------).).....)))). ( -26.60) >DroSec_CAF1 33345 112 + 1 AGA--CGUCUGAGUGUCUACUCGUCGCUCAUUCUGGAUCCGAAUCAGAAACUGAUUCAGAAUCGGAGUCAGAAUCGGAAUCGCAAUUCUAGUAUGAGUCUG------AGUAUAAGUCUGU (((--((.(.....).)((((((..((((((.(((((..(((.((.((..(((((((.......)))))))..)).)).)))....))))).)))))).))------))))...)))).. ( -34.60) >DroSim_CAF1 31552 112 + 1 AGA--CGUCUGAGUGUCUACUCGUCGCUCAUUCUGGAUCCGAAUCAGAAUCGGAUUCAGAAUCGGAGUCAGAAUCAGAAUCGCAAUUCGAGUAUGAGGCUG------AGUUUAAGUCUGU (((--(...((((((.(.....).))))))((((((((((((.......))))))))))))(..((.((((..(((...(((.....)))...)))..)))------).))..))))).. ( -40.60) >DroEre_CAF1 36209 109 + 1 -----CGUCUGAGUGGCUACUCGUCGCUCAUUCUGGAUCCGAAUCGGAC------UCCGAAUCGGAUUCAGAAUCGGAAUCAGAAUCCGAAUCGGAGUCUGAGUCUGAGUUUAAGUCUGG -----....((((((((.....))))))))((((((((((((.((((..------.)))).))))))))))))(((((.(((((.((((...)))).))))).)))))............ ( -50.40) >DroYak_CAF1 34455 108 + 1 UGCCAUGUCUGAGUGUCUACUCGUCGCUCAUUCUGGAUCCGAAUCCGAA------UCAGAAUCGGAUUCAGAAUCGGAAUCGCAAUCUGCAUCGGAGUCUG------AGUAUAAGUCUGG ..(((.(.(((((((.(.....).))))).((((((((((((.((....------...)).))))))))))))(((((.((((.....))....)).))))------).....)).)))) ( -32.50) >DroPer_CAF1 50675 90 + 1 AGC--CGUCUGAGUGUCUACUCGUCUCUGAGUCUGAAUCUGAAUCUGAA------UC-----CCGAGUCUGAGUC-----------UCGAGUCUGGGUCUG------CUUCUGCGUCUGG ((.--(((..(((((.(((((((.(((.((.((.((.((.......)).------))-----..)).)).)))..-----------.)))))...)).).)------)))..))).)).. ( -20.80) >consensus AGC__CGUCUGAGUGUCUACUCGUCGCUCAUUCUGGAUCCGAAUCAGAA______UCAGAAUCGGAGUCAGAAUCGGAAUCGCAAUCCGAGUAUGAGUCUG______AGUAUAAGUCUGG ......(.(((((((.(.....).)))))(((((((.(((((.((.((.......)).)).))))).))))))).....................)).)..................... (-15.35 = -16.78 + 1.43)

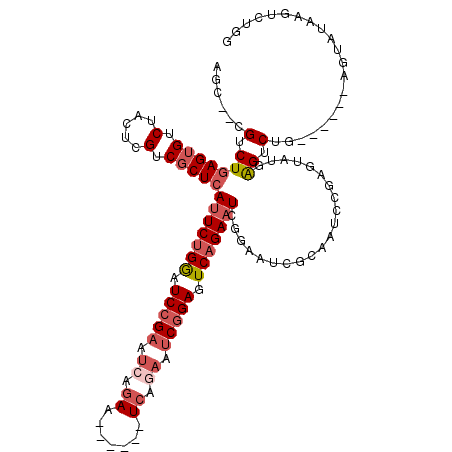
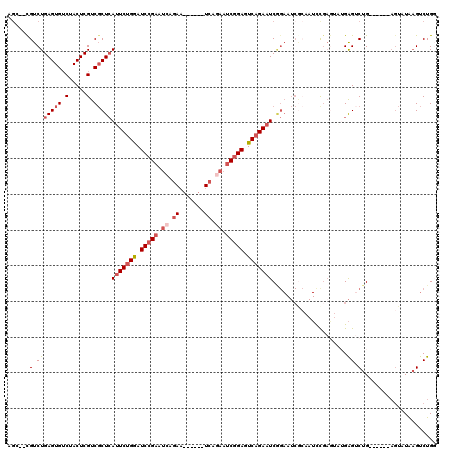
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:29 2006