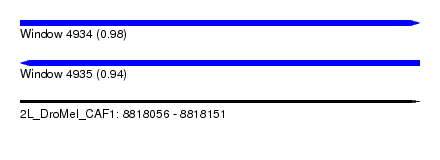
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,818,056 – 8,818,151 |
| Length | 95 |
| Max. P | 0.981051 |

| Location | 8,818,056 – 8,818,151 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 92.27 |
| Mean single sequence MFE | -22.39 |
| Consensus MFE | -21.81 |
| Energy contribution | -21.80 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8818056 95 + 22407834 GAUCUAAUUGCAGAACGGGGCAACCUUUUUCUCCUUCCCAACUCGUAUUUU-----UUUUU---UUUUUUGCCCAGCCCCGGCGACUGCAAUUAAAAUGGAAC ....(((((((((..((((((..............................-----.....---...........))))))....)))))))))......... ( -21.45) >DroSec_CAF1 26232 103 + 1 GAUCUAAUUGCAGAACGGGGCGACCUUUUUCUCCUUCCCUACUCGUAUUUUACUUAUUUUUUUUUUUUUUGCCCAGCCCCGGCGACUGCAAUUAAAAUGGAAC ....(((((((((..((((((((......)).............(((...)))......................))))))....)))))))))......... ( -22.90) >DroSim_CAF1 26024 100 + 1 GAUCUAAUUGCAGAACGGGGCGACCUUUUUCUCCUUCCCUACUCGUAUUUUACUUAUUUUU---UUUUUUGCCCAGCCCCGGCGACUGCAAUUAAAAUGGAAC ....(((((((((..((((((((......)).............(((...)))........---...........))))))....)))))))))......... ( -22.90) >DroEre_CAF1 30755 98 + 1 GAUCUAAUUGCAGAACGGGGCGACCUU-UUCUCCUCCCCUACUUGUUUUUUC-UUUUUUUU---UUUUUUGCCCAGCCCCGGCGACUGCAAUUAAAAUGGAAC ....(((((((((...((((.(.....-.....).)))).............-........---....(((((.......))))))))))))))......... ( -22.30) >consensus GAUCUAAUUGCAGAACGGGGCGACCUUUUUCUCCUUCCCUACUCGUAUUUUA_UUAUUUUU___UUUUUUGCCCAGCCCCGGCGACUGCAAUUAAAAUGGAAC ....(((((((((..((((((......................................................))))))....)))))))))......... (-21.81 = -21.80 + -0.00)

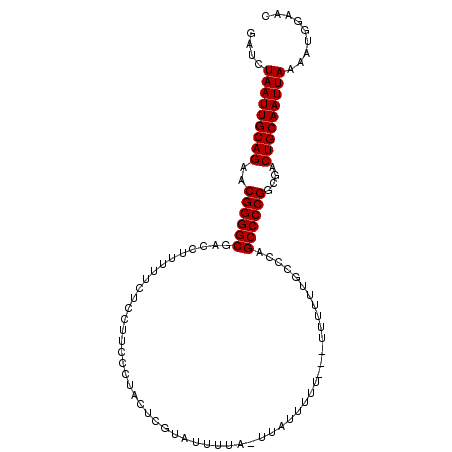
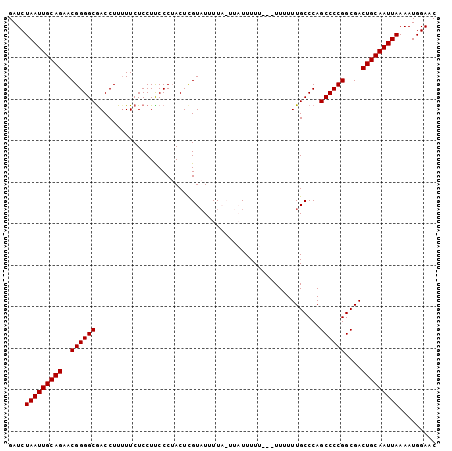
| Location | 8,818,056 – 8,818,151 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 92.27 |
| Mean single sequence MFE | -24.61 |
| Consensus MFE | -23.76 |
| Energy contribution | -23.57 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8818056 95 - 22407834 GUUCCAUUUUAAUUGCAGUCGCCGGGGCUGGGCAAAAAA---AAAAA-----AAAAUACGAGUUGGGAAGGAGAAAAAGGUUGCCCCGUUCUGCAAUUAGAUC .......(((((((((((....((((((...((......---.....-----..(((....)))...............)).))))))..))))))))))).. ( -23.39) >DroSec_CAF1 26232 103 - 1 GUUCCAUUUUAAUUGCAGUCGCCGGGGCUGGGCAAAAAAAAAAAAAAUAAGUAAAAUACGAGUAGGGAAGGAGAAAAAGGUCGCCCCGUUCUGCAAUUAGAUC .......(((((((((((....((((((..(((..............((.(((...)))...))...............)))))))))..))))))))))).. ( -25.45) >DroSim_CAF1 26024 100 - 1 GUUCCAUUUUAAUUGCAGUCGCCGGGGCUGGGCAAAAAA---AAAAAUAAGUAAAAUACGAGUAGGGAAGGAGAAAAAGGUCGCCCCGUUCUGCAAUUAGAUC .......(((((((((((....((((((...........---........(((...))).............((......))))))))..))))))))))).. ( -24.80) >DroEre_CAF1 30755 98 - 1 GUUCCAUUUUAAUUGCAGUCGCCGGGGCUGGGCAAAAAA---AAAAAAAA-GAAAAAACAAGUAGGGGAGGAGAA-AAGGUCGCCCCGUUCUGCAAUUAGAUC .......(((((((((((.((((.......)))......---........-.............((((.((....-....)).)))))..))))))))))).. ( -24.80) >consensus GUUCCAUUUUAAUUGCAGUCGCCGGGGCUGGGCAAAAAA___AAAAAUAA_UAAAAUACGAGUAGGGAAGGAGAAAAAGGUCGCCCCGUUCUGCAAUUAGAUC .......(((((((((((....((((((..(((..............................................)))))))))..))))))))))).. (-23.76 = -23.57 + -0.19)

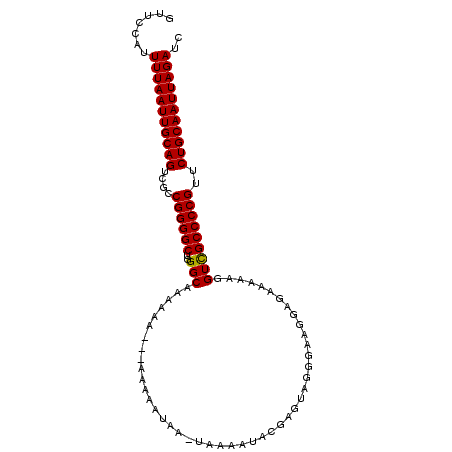
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:23 2006