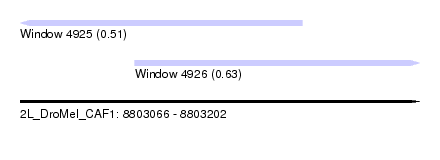
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,803,066 – 8,803,202 |
| Length | 136 |
| Max. P | 0.633108 |

| Location | 8,803,066 – 8,803,162 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 79.45 |
| Mean single sequence MFE | -20.28 |
| Consensus MFE | -16.30 |
| Energy contribution | -17.02 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8803066 96 - 22407834 CCAGCCGAACCGA-----AAUACUCGCCCAUCCAUCCAGGCGAACCUUCCGUCCUUCUGC--------CCGGCCAUUCAAGUCAUCCAUCUCGGCUUUGGCAGCUUUCG ...((((((((((-----.....(((((..........))))).....(((..(....).--------.)))..................)))).))))))........ ( -23.50) >DroSec_CAF1 11130 96 - 1 CCAGCCGAACCGA-----AAUACUCGCCCAUCCAUCCAGGCGAACCUUCUGUCCUUCUGC--------CCGGCCAUUCAAGUCAUCCAUCUCGGCUUUGGCAGCUUUCG ...((((((((((-----.....(((((..........))))).................--------..(((.......))).......)))).))))))........ ( -22.00) >DroSim_CAF1 10946 96 - 1 CCAGCCGAACCGA-----AAUACUCGCCCAUCCAUCCAGGCGAACCUUCUGUCCUUCUGC--------CCGGCCAUUCAAGUCAUCCAUCUCGGCUUUGGCAGCUUUCG ...((((((((((-----.....(((((..........))))).................--------..(((.......))).......)))).))))))........ ( -22.00) >DroEre_CAF1 12699 109 - 1 CCAGCCGAACCGAAAUACUAUACUCGCCCAUCCAUCCAGGCGAACCUUCUGUCCUUCUGUCCUCCUGCCCGGCCAUCCAAGUCAUCCAUCUCGGCUUUGGCAGCUUUCG ...((((((((((..........(((((..........)))))...........................(((.......))).......)))).))))))........ ( -22.00) >DroAna_CAF1 13292 78 - 1 CCAGCCAGGCCAA-----AA-CCUAAACCAUCCAUCCAUCCAAAU-------------------------GUCCAUCCAAGCCAUCCAUCUCGGAUUUGGCAGCUCUUG ..(((.(((....-----..-))).....................-------------------------..........(((((((.....)))..)))).))).... ( -11.90) >consensus CCAGCCGAACCGA_____AAUACUCGCCCAUCCAUCCAGGCGAACCUUCUGUCCUUCUGC________CCGGCCAUUCAAGUCAUCCAUCUCGGCUUUGGCAGCUUUCG ...((((((((((..........(((((..........)))))...........................(((.......))).......)))).))))))........ (-16.30 = -17.02 + 0.72)

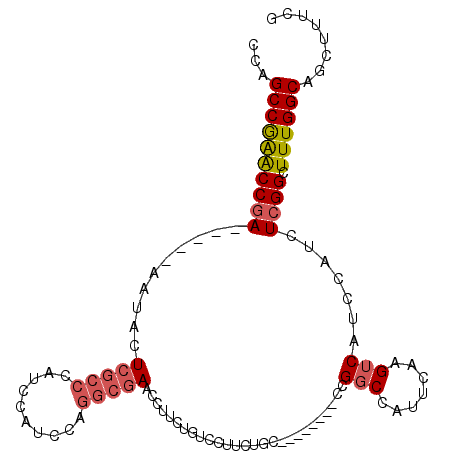

| Location | 8,803,105 – 8,803,202 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 91.63 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -25.42 |
| Energy contribution | -25.43 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.633108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8803105 97 + 22407834 GG--------GCAGAAGGACGGAAGGUUCGCCUGGAUGGAUGGGCGAGUAUU-----UCGGUUCGGCUGGAUGGGUUGUCAGUCGGUGGGCCAGCUCAUAUGAAAUUUUA ((--------((((..((((.((((..(((((((......)))))))...))-----)).))))..))...(((.(..((....))..).)))))))............. ( -32.50) >DroSec_CAF1 11169 97 + 1 GG--------GCAGAAGGACAGAAGGUUCGCCUGGAUGGAUGGGCGAGUAUU-----UCGGUUCGGCUGGAUGGGUUGUCAGUUGGUGGGCCAGCUCAUAUGAAAUUUUA ((--------((((..((((.((((..(((((((......)))))))...))-----)).))))..))...(((.(..((....))..).)))))))............. ( -31.00) >DroSim_CAF1 10985 97 + 1 GG--------GCAGAAGGACAGAAGGUUCGCCUGGAUGGAUGGGCGAGUAUU-----UCGGUUCGGCUGGAUGGGUUGUCAGUUGGUGGGCCAGCUCAUAUGAAAUUUUA ((--------((((..((((.((((..(((((((......)))))))...))-----)).))))..))...(((.(..((....))..).)))))))............. ( -31.00) >DroEre_CAF1 12738 110 + 1 GGGCAGGAGGACAGAAGGACAGAAGGUUCGCCUGGAUGGAUGGGCGAGUAUAGUAUUUCGGUUCGGCUGGAUGGGUUGUCAGUCGGUGGGCCAGCUCAUAUGAAAUUUUA ((((....(..(....)..)....((((((((.((((.((...((.......))...)).))))((((((........)))))))))))))).))))............. ( -33.10) >consensus GG________GCAGAAGGACAGAAGGUUCGCCUGGAUGGAUGGGCGAGUAUU_____UCGGUUCGGCUGGAUGGGUUGUCAGUCGGUGGGCCAGCUCAUAUGAAAUUUUA ...........(((..((((.(((..((((((((......))))))))..)......)).))))..))).((((((((((........)).))))))))........... (-25.42 = -25.43 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:14 2006