| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
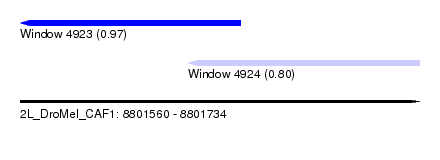
| Location | 8,801,560 – 8,801,734 |
| Length | 174 |
| Max. P | 0.972293 |

| Location | 8,801,560 – 8,801,656 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 77.39 |
| Mean single sequence MFE | -41.40 |
| Consensus MFE | -23.29 |
| Energy contribution | -26.29 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
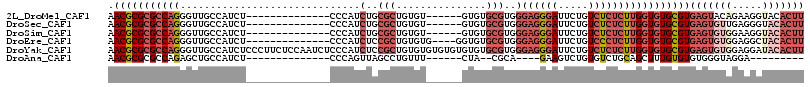
>2L_DroMel_CAF1 8801560 96 - 22407834 AACGCGCGCCAGGGUUGCCAUCU--------------CCCAUCUGCGCUGUGU------GUGUGCGUGGGAGGGAUUCUGUCUCUCUUGGUGUGCGUGAGUACAGAAGGUACACUU .(((((((((((((..(.(((((--------------(((((.(((((.....------))))).)))))))).....)).)..)))))))))))))..((((.....)))).... ( -42.00) >DroSec_CAF1 9388 96 - 1 AACGCGCGCCAGGGUUGCCAUCU--------------CCCAUCUGCGCUGUGU------GUGUGCGUGGGAGGGAUUCUGUCUCUCUUGGUGUGCGUGAGUGUUGAGGGUACACUU .(((((((((((((..(.(((((--------------(((((.(((((.....------))))).)))))))).....)).)..)))))))))))))((((((.......)))))) ( -43.10) >DroSim_CAF1 9406 96 - 1 AACGCGCGCCAGGGUUGCCAUCU--------------CCCAUCUGCGCUGUGU------GUGUGCGUGGGAGGGAUUCUGUCUCUCUUGGUGUGCGUGAGUGUGGAAGGUACACUU .(((((((((((((..(.(((((--------------(((((.(((((.....------))))).)))))))).....)).)..)))))))))))))(((((((.....))))))) ( -43.90) >DroEre_CAF1 11070 98 - 1 AACGCGCGCCAGGGUUGCCAUCU--------------CCCAUCUCCGCUGUGUG----GGUGUGCGUGGGAGGGAUUCUGUCCCUCUUGGUGUGCGUGAGUGUGGAGGCUACACUU .(((((((((((((........(--------------(((((...((((.....----))))...))))))(((((...))))))))))))))))))((((((((...)))))))) ( -45.00) >DroYak_CAF1 11348 116 - 1 AACGCGCGCCAGGGUUGCCAUCUCCCUUCUCCAAUCUCCCAUCUCCGCUGUGUGUGUGUGUGUGCGUGGGAGGGAUUCUGUCUCUCUUGGUGUGCGUGAGUGUGGAGGAUACACUU .(((((((((((((..(.((..(((((((..(.............(((.....))).((....)))..)))))))...)).)..)))))))))))))(((((((.....))))))) ( -42.30) >DroAna_CAF1 11874 81 - 1 AACGCGCGCCAGAGCUGCCAUCU--------------CCCAGUUAGCCUGUUU------CUA--CGCA----GAAGUCUGUGUCUGCAGCUUUGUGUGUGGGUAGGA--------- .((.((((((((((((((.....--------------..(((.....)))...------..(--((((----(....))))))..))))))))).))))).))....--------- ( -32.10) >consensus AACGCGCGCCAGGGUUGCCAUCU______________CCCAUCUGCGCUGUGU______GUGUGCGUGGGAGGGAUUCUGUCUCUCUUGGUGUGCGUGAGUGUGGAAGGUACACUU .(((((((((((..............................(..(((...............)))..)((((((.....)))))))))))))))))(((((((.....))))))) (-23.29 = -26.29 + 3.00)
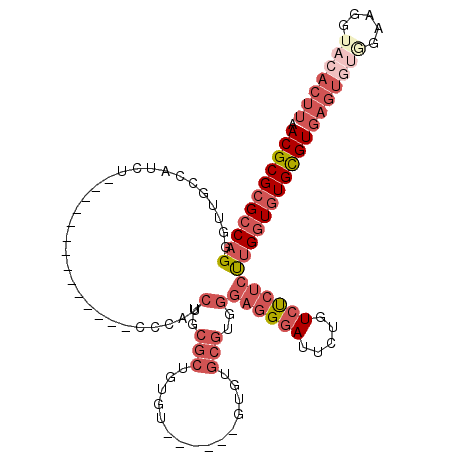
| Location | 8,801,633 – 8,801,734 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.15 |
| Mean single sequence MFE | -29.37 |
| Consensus MFE | -21.98 |
| Energy contribution | -22.28 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8801633 101 - 22407834 ACUGUCGGGUAAAAGCAGCCAGAAACUCUUGGCAA-AAAGGAGUUGGCUAACAUAAUUGAA-AAGCGCUUUUCUAUUAAGAACGCGCGCCAGGGU-UGCC---AUCU------------- ..............((((((...(((((((.....-..)))))))(((...((....))..-..((((..((((....)))).)))))))..)))-))).---....------------- ( -29.20) >DroPse_CAF1 22823 100 - 1 AGAAUCAGGCAGGAGCAG----GAACA---GGCAGCAAAGGAGUUGGCUAGCAUAAUUGAAAAAGCGCUUUUCUAUUAAGAACGCGCGCCAGGGUAUGCCUCAGUCU------------- (((...(((((...((.(----(...(---(.((((......)))).))...............((((..((((....)))).)))).))...)).)))))...)))------------- ( -24.90) >DroEre_CAF1 11145 101 - 1 ACUGACGGGUAAAAGCAGCCAGAAACUCUUGGCAA-AAAGGAGUUGGCUAACAUAAUUGAA-AAGCGCUUUUCUAUUAAGAACGCGCGCCAGGGU-UGCC---AUCU------------- ..............((((((...(((((((.....-..)))))))(((...((....))..-..((((..((((....)))).)))))))..)))-))).---....------------- ( -29.20) >DroYak_CAF1 11428 114 - 1 ACUGCCUGGUGAAAGCAGCCAGAAACUCUUGGCAA-AAAGGAGUUGGCUAACAUAAUUGAA-AAGCGCUUUUCUAUUAAGAACGCGCGCCAGGGU-UGCC---AUCUCCCUUCUCCAAUC ......(((.(((....(((((......)))))..-...((((.((((.(((....(((..-..((((..((((....)))).))))..))).))-))))---).)))).))).)))... ( -35.10) >DroAna_CAF1 11932 101 - 1 GCAGGGAGGCAAAUGCUGCCAGAAACUCUUGGCAA-AAAGGAGUUGGCCAACAUAAUUGAA-AAGCGCUUUUCUAUUAAGAACGCGCGCCAGAGC-UGCC---AUCU------------- ((((...((((.....)))).....((((.(((..-...((......))............-..((((..((((....)))).))))))))))))-))).---....------------- ( -30.30) >DroPer_CAF1 23038 100 - 1 AGAUUCAGGCAGGAGCAG----GAACA---GGCAGCAAAGGAGUUGGCUAGCAUAAUUGAAAAAGCGCUUUUCUAUUAAGAACGCGCGCCAGGGUAUGCCUCAGUCU------------- (((((.(((((...((.(----(...(---(.((((......)))).))...............((((..((((....)))).)))).))...)).))))).)))))------------- ( -27.50) >consensus ACAGUCAGGCAAAAGCAGCCAGAAACUCUUGGCAA_AAAGGAGUUGGCUAACAUAAUUGAA_AAGCGCUUUUCUAUUAAGAACGCGCGCCAGGGU_UGCC___AUCU_____________ .......((((...((.(((((......)))))..........(((((...((....)).....((((..((((....)))).))))))))).)).)))).................... (-21.98 = -22.28 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:13 2006