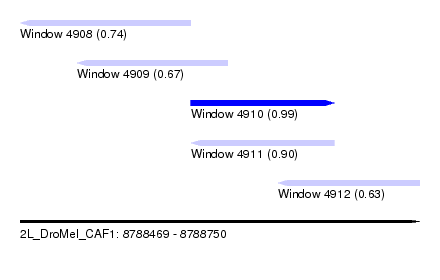
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,788,469 – 8,788,750 |
| Length | 281 |
| Max. P | 0.994249 |

| Location | 8,788,469 – 8,788,589 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.58 |
| Mean single sequence MFE | -41.57 |
| Consensus MFE | -37.38 |
| Energy contribution | -37.13 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
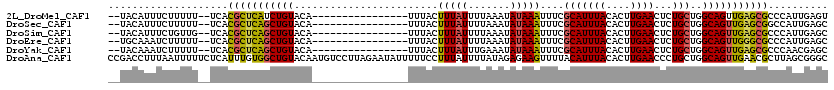
>2L_DroMel_CAF1 8788469 120 - 22407834 GACAUCGGCGGGAACGUCAUCGUUGAGGCACAAGAACAAAAGGACUUCCGGUGACGUUUAAAUGCGUGGCAUGUCACAGCGCAGCGGCAGACAGCAGCAUCCUGGCUGAAGGCUGCCUUC ((((((.(((.(((((((((((..((((..(..........)..))))))))))))))).....))).).)))))..........(((((....((((......))))....)))))... ( -43.20) >DroSec_CAF1 91225 120 - 1 GACAUCGACGGGAACGUCAUCGUUGAGGCACAAGAACAAAAGGACUUCCGGUGACGUUUAAAUGCGUGGCAUGUCACAGCGCAGCGGCAGACAGCAGCAUCCUGGCUGAAGGCUGCCUUC ((((((.(((.(((((((((((..((((..(..........)..))))))))))))))).....))).).)))))..........(((((....((((......))))....)))))... ( -42.90) >DroSim_CAF1 94785 120 - 1 GACAUCGGCGGGAACGUCAUCGCUGAGGCACAAGAACAAAAGGACUUCCGGUGACGUUUAAAUGCGUGGCAUGUCACAGCGCAGCGGCAGACAGCAGCAUCCUGGCUGAAGGCUGCCUUC ((((((.(((.((((((((((((....))............((....)))))))))))).....))).).)))))..........(((((....((((......))))....)))))... ( -43.40) >DroEre_CAF1 97706 120 - 1 GACAUCGGCGGGAACGUCAUCGUUGAGGCACAAGAACAAAAGGACUUCCGGUGACGUUUAAAUGCGUGGCAUGUCACGGCGCAGCGGCAGACAGCAGCAUCCUGGCUGAAGGCCGCCUUU .......(((.(((((((((((..((((..(..........)..))))))))))))))).....(((((....))))).))).(((((...((((((....)).))))...))))).... ( -43.20) >DroYak_CAF1 96497 112 - 1 GACUUCGGCGGGAACGUCAUCGUUGAGGCACAAGAACAAAAGGACUUCCGGUGACGUUUAAAUGCGUGGCAUGUCACAGCGCAGCGGCAGACAGCAGCAUCCUGGCUGAAGG-------- ..(((((((.((((((((((((..((((..(..........)..))))))))))))))...((((...((.((((...((......)).)))))).)))).)).))))))).-------- ( -40.90) >DroAna_CAF1 113518 108 - 1 GACAUCGGCG-GAACGUCAUCGUUGAGGCAUAAGAACAAAAGGACUUCCGGUGACGUUUAAAUGCGUGGCAUGUCACAGCGCAGCGGCAGACAGCAGCAUCCUGGCUGA----------- ((((((.(((-(((((((((((..(((((.............).))))))))))))))).....))).).))))).((((.(((..((........))...))))))).----------- ( -35.82) >consensus GACAUCGGCGGGAACGUCAUCGUUGAGGCACAAGAACAAAAGGACUUCCGGUGACGUUUAAAUGCGUGGCAUGUCACAGCGCAGCGGCAGACAGCAGCAUCCUGGCUGAAGGCUGCCUUC ((((((.(((.(((((((((((..((((..(..........)..))))))))))))))).....))).).))))).((((.(((..((........))...)))))))............ (-37.38 = -37.13 + -0.25)



| Location | 8,788,509 – 8,788,615 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.37 |
| Mean single sequence MFE | -35.52 |
| Consensus MFE | -32.49 |
| Energy contribution | -32.68 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8788509 106 - 22407834 UC--------------CUGGCAUCGCCUGUCAGUCAGUUAGACAUCGGCGGGAACGUCAUCGUUGAGGCACAAGAACAAAAGGACUUCCGGUGACGUUUAAAUGCGUGGCAUGUCACAGC ..--------------.((((((.(((.....(((.....)))....(((.(((((((((((..((((..(..........)..)))))))))))))))...)))..))))))))).... ( -34.90) >DroSec_CAF1 91265 106 - 1 UC--------------CUGGCAUCGCCUGUCAGUCAGUUAGACAUCGACGGGAACGUCAUCGUUGAGGCACAAGAACAAAAGGACUUCCGGUGACGUUUAAAUGCGUGGCAUGUCACAGC ..--------------...((((..((((((.(((.....)))...))))))((((((((((..((((..(..........)..))))))))))))))...))))((((....))))... ( -33.40) >DroSim_CAF1 94825 106 - 1 UC--------------CUGGCAUCGCCUGUCAGUCAGUUAGACAUCGGCGGGAACGUCAUCGCUGAGGCACAAGAACAAAAGGACUUCCGGUGACGUUUAAAUGCGUGGCAUGUCACAGC ..--------------.((((((.(((.....(((.....)))....(((.((((((((((((....))............((....))))))))))))...)))..))))))))).... ( -35.10) >DroEre_CAF1 97746 106 - 1 UC--------------CUGGCAUCGCCUGUCAGUCAGUUAGACAUCGGCGGGAACGUCAUCGUUGAGGCACAAGAACAAAAGGACUUCCGGUGACGUUUAAAUGCGUGGCAUGUCACGGC .(--------------(((((((.(((.....(((.....)))....(((.(((((((((((..((((..(..........)..)))))))))))))))...)))..))))))))).)). ( -36.90) >DroYak_CAF1 96529 106 - 1 UC--------------CUGGCAUCGCCUGUCAGUCAGUUAGACUUCGGCGGGAACGUCAUCGUUGAGGCACAAGAACAAAAGGACUUCCGGUGACGUUUAAAUGCGUGGCAUGUCACAGC ..--------------.((((((.(((....((((.....))))...(((.(((((((((((..((((..(..........)..)))))))))))))))...)))..))))))))).... ( -35.00) >DroAna_CAF1 113547 119 - 1 UCUCCACUCACGCGUUCUGCCAUCGCCUGUCAGUCAGCUAGACAUCGGCG-GAACGUCAUCGUUGAGGCAUAAGAACAAAAGGACUUCCGGUGACGUUUAAAUGCGUGGCAUGUCACAGC .(((.((....((((((((((......((((((....)).))))..))))-))))))....)).)))((((..((((....((....))(....)))))..))))((((....))))... ( -37.80) >consensus UC______________CUGGCAUCGCCUGUCAGUCAGUUAGACAUCGGCGGGAACGUCAUCGUUGAGGCACAAGAACAAAAGGACUUCCGGUGACGUUUAAAUGCGUGGCAUGUCACAGC .................((((((.(((.....(((.....)))....(((.(((((((((((..((((..(..........)..)))))))))))))))...)))..))))))))).... (-32.49 = -32.68 + 0.19)

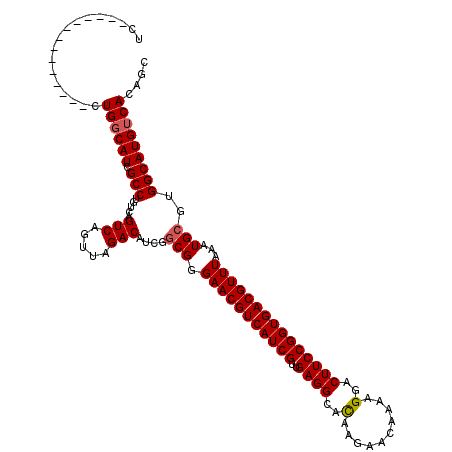
| Location | 8,788,589 – 8,788,690 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 93.90 |
| Mean single sequence MFE | -31.36 |
| Consensus MFE | -29.88 |
| Energy contribution | -30.36 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8788589 101 + 22407834 UAACUGACUGACAGGCGAUGCCAGGAGGUCCUUCUCCUGCCCCACAAAA-ACC----CCCUGCCCAACUCAAUGGGCGCUCAACUGCCAGCAGAGUUCAAGUGUAA ...(((.(((.(((..((.(((((((((....)))))))..........-...----....(((((......)))))))))..))).))))))............. ( -32.30) >DroSec_CAF1 91345 101 + 1 UAACUGACUGACAGGCGAUGCCAGGAGGUCCUUCUCCUGCCCCACAAAA-CCC----CCCUGCCCAGCUCAAUGGCCGCUCAACUGCCAGCAGAGUUCAAGUGUAA ...(((.(((.(((..((.(((((((((....)))))))..........-...----....(.(((......))).)))))..))).))))))............. ( -26.00) >DroSim_CAF1 94905 101 + 1 UAACUGACUGACAGGCGAUGCCAGGAGGUCCUUCUCCUGCCCCACAAAA-ACC----CCCUGCCCAGCUCAAUGGGCGCUCAACUGCCAGCAGAGUUCAAGUGUAA ...(((.(((.(((..((.(((((((((....)))))))..........-...----....(((((......)))))))))..))).))))))............. ( -33.10) >DroEre_CAF1 97826 105 + 1 UAACUGACUGACAGGCGAUGCCAGGAGGUCCU-CUCCUGCCCCACAGAAAGCCCCACCCCUGCCCAGCUCAAUGGGCGCCCAACUGCCAGCAGAGUUCAAGUGUAA ...(((.(((.(((.....((((((((.....-))))))......................(((((......)))))))....))).))))))............. ( -29.30) >DroYak_CAF1 96609 102 + 1 UAACUGACUGACAGGCGAUGCCAGGAGGUCCUCCUCCUGCCCCACACAAAACC----CCCUGCCCAGCUCGUUGGGCGCUCAACUGCCAGCAGAGUUCAAGUGUAA ...(((.(((.(((..((.(((((((((....)))))))..............----....((((((....))))))))))..))).))))))............. ( -36.10) >consensus UAACUGACUGACAGGCGAUGCCAGGAGGUCCUUCUCCUGCCCCACAAAA_ACC____CCCUGCCCAGCUCAAUGGGCGCUCAACUGCCAGCAGAGUUCAAGUGUAA ...(((.(((.(((..((.(((((((((....)))))))......................(((((......)))))))))..))).))))))............. (-29.88 = -30.36 + 0.48)

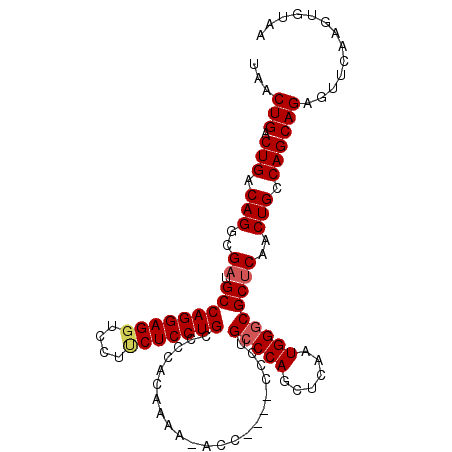
| Location | 8,788,589 – 8,788,690 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 93.90 |
| Mean single sequence MFE | -40.22 |
| Consensus MFE | -35.72 |
| Energy contribution | -35.76 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8788589 101 - 22407834 UUACACUUGAACUCUGCUGGCAGUUGAGCGCCCAUUGAGUUGGGCAGGG----GGU-UUUUGUGGGGCAGGAGAAGGACCUCCUGGCAUCGCCUGUCAGUCAGUUA .........((((..((((((((.((((((((((......)))))((((----(((-((((.(........).))))))))))).)).))).)))))))).)))). ( -42.10) >DroSec_CAF1 91345 101 - 1 UUACACUUGAACUCUGCUGGCAGUUGAGCGGCCAUUGAGCUGGGCAGGG----GGG-UUUUGUGGGGCAGGAGAAGGACCUCCUGGCAUCGCCUGUCAGUCAGUUA .........((((..((((((((.(((.((((......))))(.(((((----((.-((((.((...)).))))....))))))).).))).)))))))).)))). ( -36.10) >DroSim_CAF1 94905 101 - 1 UUACACUUGAACUCUGCUGGCAGUUGAGCGCCCAUUGAGCUGGGCAGGG----GGU-UUUUGUGGGGCAGGAGAAGGACCUCCUGGCAUCGCCUGUCAGUCAGUUA .........((((..((((((((.((((((((((......)))))((((----(((-((((.(........).))))))))))).)).))).)))))))).)))). ( -41.30) >DroEre_CAF1 97826 105 - 1 UUACACUUGAACUCUGCUGGCAGUUGGGCGCCCAUUGAGCUGGGCAGGGGUGGGGCUUUCUGUGGGGCAGGAG-AGGACCUCCUGGCAUCGCCUGUCAGUCAGUUA .........((((..((((((((.(((((.(((((.(((((..((....))..)))))...))))).((((((-.....)))))))).))).)))))))).)))). ( -41.60) >DroYak_CAF1 96609 102 - 1 UUACACUUGAACUCUGCUGGCAGUUGAGCGCCCAACGAGCUGGGCAGGG----GGUUUUGUGUGGGGCAGGAGGAGGACCUCCUGGCAUCGCCUGUCAGUCAGUUA .........((((..((((((((.((((((((((......)))))....----..............(((((((....))))))))).))).)))))))).)))). ( -40.00) >consensus UUACACUUGAACUCUGCUGGCAGUUGAGCGCCCAUUGAGCUGGGCAGGG____GGU_UUUUGUGGGGCAGGAGAAGGACCUCCUGGCAUCGCCUGUCAGUCAGUUA .........((((..((((((((.((((((((((......)))))......................((((((......)))))))).))).)))))))).)))). (-35.72 = -35.76 + 0.04)



| Location | 8,788,650 – 8,788,750 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.31 |
| Mean single sequence MFE | -20.22 |
| Consensus MFE | -14.96 |
| Energy contribution | -14.85 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8788650 100 - 22407834 --UACAUUUCUUUUU--UCACGCUCAUCUGUACA----------------UUUACUUUAUUUUAAAUAUAAAUUUCGCAUUUACACUUGAACUCUGCUGGCAGUUGAGCGCCCAUUGAGU --.......(((...--...((((((.((((..(----------------((((........))))).........(((((((....))))...)))..)))).))))))......))). ( -15.40) >DroSec_CAF1 91406 100 - 1 --UACAUUUCUUUUU--UCACGCUCAGCUGUACA----------------UUUACUUUAUUUUAAAUAUAAAUUUCGCAUUUACACUUGAACUCUGCUGGCAGUUGAGCGGCCAUUGAGC --.......(((...--...(((((((((((..(----------------((((........))))).........(((((((....))))...)))..)))))))))))......))). ( -20.40) >DroSim_CAF1 94966 100 - 1 --UACAUUUCUGUUG--UCACGCUCAGCUGUACA----------------UUUACUUUAUUUUAAAUAUAAAUUUCGCAUUUACACUUGAACUCUGCUGGCAGUUGAGCGCCCAUUGAGC --.....(((...((--...(((((((((((..(----------------((((........))))).........(((((((....))))...)))..)))))))))))..))..))). ( -19.50) >DroEre_CAF1 97891 100 - 1 --UGCAAAUCUUUUU--UCACGCUCAGCUGUACA----------------UUUACUUUAUUUUAAAUAUAAAUUUCGCAUUUACACUUGAACUCUGCUGGCAGUUGGGCGCCCAUUGAGC --.......(((...--...(((((((((((..(----------------((((........))))).........(((((((....))))...)))..)))))))))))......))). ( -18.40) >DroYak_CAF1 96671 100 - 1 --UACAAAUCUUUUU--UCACGCUCAGCUGUACA----------------UUUACUUUAUUUGAAAUAUAAAUUUCGCAUUUACACUUGAACUCUGCUGGCAGUUGAGCGCCCAACGAGC --.............--...(((((((((((...----------------............(((((....)))))(((((((....))))...)))..))))))))))).......... ( -21.60) >DroAna_CAF1 113681 120 - 1 CCGACCUUUAAUUUUUCUCAUUUGUGGCUGUACAAUGUCCUUAGAAUAUUUUUCCUUUAUUUUAUAGAGAAGUUUUACAUUUACACUUGAACCCUGCUGGCAGUUGAACGCUUAGCGGGC .................(((..(((((.((((.(((.(((((((((((.........))))))).)).)).))).)))).)))))..)))..(((((((((........)).))))))). ( -26.00) >consensus __UACAUUUCUUUUU__UCACGCUCAGCUGUACA________________UUUACUUUAUUUUAAAUAUAAAUUUCGCAUUUACACUUGAACUCUGCUGGCAGUUGAGCGCCCAUUGAGC ....................(((((((((((........................(((((.......)))))....(((((((....))))...)))..))))))))))).......... (-14.96 = -14.85 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:01 2006