
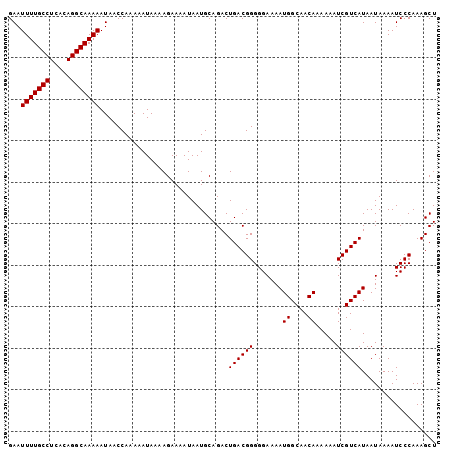
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,786,304 – 8,786,433 |
| Length | 129 |
| Max. P | 0.997028 |

| Location | 8,786,304 – 8,786,407 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 99.35 |
| Mean single sequence MFE | -16.40 |
| Consensus MFE | -16.40 |
| Energy contribution | -16.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8786304 103 - 22407834 GAAUUUUGCCUCACAGGCAAAAAUAACCAAAAAUAAAAGAAAAUAAUGCAGACUGACGGGGGAAAAUGGCAACAAAAAAUCGUCAUAAUAAAAUCCCAAAGCU ...(((((((.....)))))))...............................((((((.......((....)).....)))))).................. ( -16.40) >DroSec_CAF1 89014 103 - 1 GAAUUUUGCCUGACAGGCAAAAAUAACCAAAAAUAAAAGAAAAUAAUGCAGACUGACGGGGGAAAAUGGCAACAAAAAAUCGUCAUAAUAAAAUCCCAAAGCU ...(((((((.....)))))))...............................((((((.......((....)).....)))))).................. ( -16.40) >DroSim_CAF1 92538 103 - 1 GAAUUUUGCCUCACAGGCAAAAAUAACCAAAAAUAAAAGAAAAUAAUGCAGACUGACGGGGGAAAAUGGCAACAAAAAAUCGUCAUAAUAAAAUCCCAAAGCU ...(((((((.....)))))))...............................((((((.......((....)).....)))))).................. ( -16.40) >consensus GAAUUUUGCCUCACAGGCAAAAAUAACCAAAAAUAAAAGAAAAUAAUGCAGACUGACGGGGGAAAAUGGCAACAAAAAAUCGUCAUAAUAAAAUCCCAAAGCU ...(((((((.....)))))))...............................((((((.......((....)).....)))))).................. (-16.40 = -16.40 + 0.00)


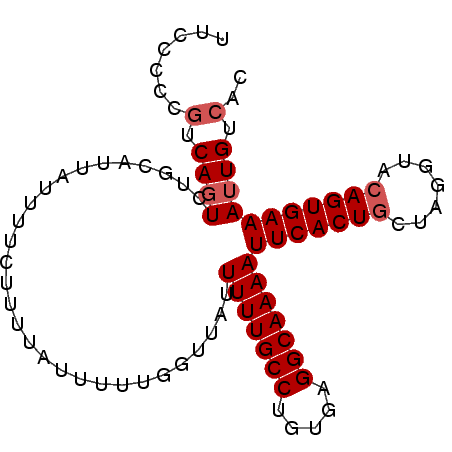
| Location | 8,786,343 – 8,786,433 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 94.89 |
| Mean single sequence MFE | -17.42 |
| Consensus MFE | -15.10 |
| Energy contribution | -15.70 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8786343 90 + 22407834 UUCCCCCGUCAGUCUGCAUUAUUUUCUUUUAUUUUUGGUUAUUUUUGCCUGUGAGGCAAAAUUCACUGCUAGGUACAGUGAAAUUGUCAC .......(.((((.............................(((((((.....)))))))(((((((.......))))))))))).).. ( -17.60) >DroSec_CAF1 89053 90 + 1 UUCCCCCGUCAGUCUGCAUUAUUUUCUUUUAUUUUUGGUUAUUUUUGCCUGUCAGGCAAAAUUCACUGCUAGGUACAGUGAAAUUGUCAC .......(.((((.............................(((((((.....)))))))(((((((.......))))))))))).).. ( -17.60) >DroSim_CAF1 92577 90 + 1 UUCCCCCGUCAGUCUGCAUUAUUUUCUUUUAUUUUUGGUUAUUUUUGCCUGUGAGGCAAAAUUCACUGCUAGGUACAGUGAAAUUGUCAC .......(.((((.............................(((((((.....)))))))(((((((.......))))))))))).).. ( -17.60) >DroEre_CAF1 95603 87 + 1 UUUCCCCGUCAUUCUGCAUUAUUUUCUUUUAUUUUCGGUUAUUUUUGCCUGUGAGGCAAAAUUCACU---ACGUACAGUGAAAUUGUGAC .......(((((..............................(((((((.....)))))))((((((---......))))))...))))) ( -16.70) >DroYak_CAF1 94264 90 + 1 UUUUCCCGUCAGUCUGCAUUAUUUUCCUUUAUUUUUGGUUAUUUUUGCCUGUGAGGCAAAAUUCACUGCUAGGUACAGUGAAAUUGUCAC .......(.((((.............................(((((((.....)))))))(((((((.......))))))))))).).. ( -17.60) >consensus UUCCCCCGUCAGUCUGCAUUAUUUUCUUUUAUUUUUGGUUAUUUUUGCCUGUGAGGCAAAAUUCACUGCUAGGUACAGUGAAAUUGUCAC .......(.((((.............................(((((((.....)))))))(((((((.......))))))))))).).. (-15.10 = -15.70 + 0.60)


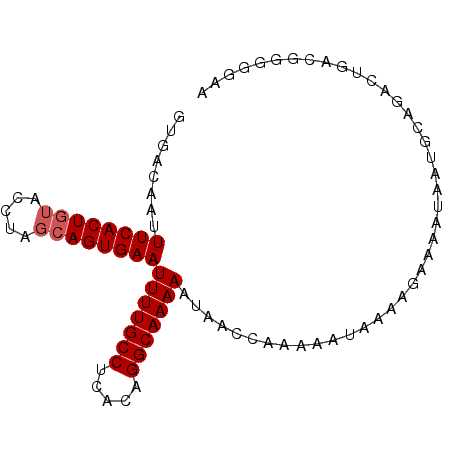
| Location | 8,786,343 – 8,786,433 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 94.89 |
| Mean single sequence MFE | -18.30 |
| Consensus MFE | -16.28 |
| Energy contribution | -16.68 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.997028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8786343 90 - 22407834 GUGACAAUUUCACUGUACCUAGCAGUGAAUUUUGCCUCACAGGCAAAAAUAACCAAAAAUAAAAGAAAAUAAUGCAGACUGACGGGGGAA .((.((..((((((((.....))))))))(((((((.....)))))))........................)))).............. ( -18.00) >DroSec_CAF1 89053 90 - 1 GUGACAAUUUCACUGUACCUAGCAGUGAAUUUUGCCUGACAGGCAAAAAUAACCAAAAAUAAAAGAAAAUAAUGCAGACUGACGGGGGAA .((.((..((((((((.....))))))))(((((((.....)))))))........................)))).............. ( -18.00) >DroSim_CAF1 92577 90 - 1 GUGACAAUUUCACUGUACCUAGCAGUGAAUUUUGCCUCACAGGCAAAAAUAACCAAAAAUAAAAGAAAAUAAUGCAGACUGACGGGGGAA .((.((..((((((((.....))))))))(((((((.....)))))))........................)))).............. ( -18.00) >DroEre_CAF1 95603 87 - 1 GUCACAAUUUCACUGUACGU---AGUGAAUUUUGCCUCACAGGCAAAAAUAACCGAAAAUAAAAGAAAAUAAUGCAGAAUGACGGGGAAA ((((....(((((((....)---))))))(((((((.....)))))))...............................))))....... ( -18.10) >DroYak_CAF1 94264 90 - 1 GUGACAAUUUCACUGUACCUAGCAGUGAAUUUUGCCUCACAGGCAAAAAUAACCAAAAAUAAAGGAAAAUAAUGCAGACUGACGGGAAAA ((((.....))))....(((..((((...(((((((.....)))))))....((.........))............))))..))).... ( -19.40) >consensus GUGACAAUUUCACUGUACCUAGCAGUGAAUUUUGCCUCACAGGCAAAAAUAACCAAAAAUAAAAGAAAAUAAUGCAGACUGACGGGGGAA ........((((((((.....))))))))(((((((.....))))))).......................................... (-16.28 = -16.68 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:55 2006