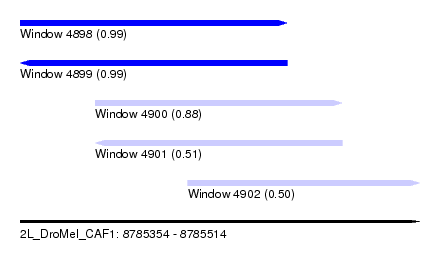
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,785,354 – 8,785,514 |
| Length | 160 |
| Max. P | 0.988561 |

| Location | 8,785,354 – 8,785,461 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.56 |
| Mean single sequence MFE | -28.37 |
| Consensus MFE | -24.67 |
| Energy contribution | -24.83 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8785354 107 + 22407834 ---U--UUUCUGUCACCUGUUUCAACACUUGAAAU--------GUGACGGCAGCCGCCAAACAACCUAAUUAGUAUGCAAAACACAUUAAGUGGCAGCCAACUACGUUGCCGCAGUACUC ---.--.....(((((..((((((.....))))))--------)))))(((....)))...........(((((.((.....)).)))))((((((((.......))))))))....... ( -27.70) >DroSec_CAF1 88067 108 + 1 U--U--UUUCUGUCACCUGUUUCAACACUUGAAAU--------GUGACGGCAGCCGCCAAACAACCUAAUUAGUGUGCAAAACACAUUAAGUGGCAGCCAACUACGUUGCCGCAGUACUC .--.--.....(((((..((((((.....))))))--------)))))(((....)))...........((((((((.....))))))))((((((((.......))))))))....... ( -33.70) >DroSim_CAF1 91582 110 + 1 UUUU--UUUCUGUCACCUGUUUCAACACUUGAAAU--------GUGACGGCAGCCGCCAAACAACCUAAUUAGUAUGCAAAACACAUUAAGUGGCAGCCAACUACGUUGCCGCAGUACUC ....--.....(((((..((((((.....))))))--------)))))(((....)))...........(((((.((.....)).)))))((((((((.......))))))))....... ( -27.70) >DroEre_CAF1 94587 112 + 1 UUUUUAUUUCUGUCACCUGUUUCAACACUUGAAAU--------GUGACGGCAGCCGCCAAACAACCUAAUUAGUAUGCAAAACACAUUAAGUGGCAGCCAACUACGUUGCCGCAGUACUC ...........(((((..((((((.....))))))--------)))))(((....)))...........(((((.((.....)).)))))((((((((.......))))))))....... ( -27.70) >DroYak_CAF1 93200 107 + 1 UU-----UUCUGUCACCUGUUUCAACACUUGAAAU--------GUGACGGCAGCCGCCAAACAACCUAAUUAGUAUGCAAAACACAUUAAGUGGCAGCCAACUACGUUGCCGCAGUACUC ..-----....(((((..((((((.....))))))--------)))))(((....)))...........(((((.((.....)).)))))((((((((.......))))))))....... ( -27.70) >DroAna_CAF1 110226 116 + 1 U--U--UUUCUGUCACCUGUUCCAACACUUGAAAUGUGAAAAUGUGACGGCAGCCGCCAAACAACCUAAUUAGUAUGCAAAACACAUUAAGUGGCAGCCAACUACGUUGCCGCAGUACUC .--.--.....(((((...(((((..........)).)))...)))))(((....)))...........(((((.((.....)).)))))((((((((.......))))))))....... ( -25.70) >consensus U__U__UUUCUGUCACCUGUUUCAACACUUGAAAU________GUGACGGCAGCCGCCAAACAACCUAAUUAGUAUGCAAAACACAUUAAGUGGCAGCCAACUACGUUGCCGCAGUACUC ...........(((((..((((((.....))))))........)))))(((....)))...........(((((.((.....)).)))))((((((((.......))))))))....... (-24.67 = -24.83 + 0.17)

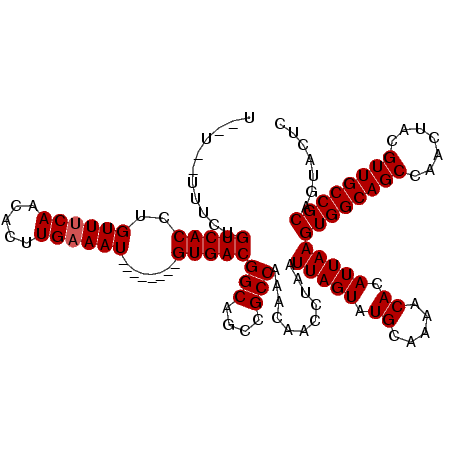
| Location | 8,785,354 – 8,785,461 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.56 |
| Mean single sequence MFE | -35.42 |
| Consensus MFE | -31.09 |
| Energy contribution | -30.82 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8785354 107 - 22407834 GAGUACUGCGGCAACGUAGUUGGCUGCCACUUAAUGUGUUUUGCAUACUAAUUAGGUUGUUUGGCGGCUGCCGUCAC--------AUUUCAAGUGUUGAAACAGGUGACAGAAA--A--- ....((((((....)))))).((((((((..((((.((.((........)).)).))))..))))))))...(((((--------.(((((.....)))))...))))).....--.--- ( -34.80) >DroSec_CAF1 88067 108 - 1 GAGUACUGCGGCAACGUAGUUGGCUGCCACUUAAUGUGUUUUGCACACUAAUUAGGUUGUUUGGCGGCUGCCGUCAC--------AUUUCAAGUGUUGAAACAGGUGACAGAAA--A--A ....((((((....)))))).((((((((..(((((((.....))).((....))))))..))))))))...(((((--------.(((((.....)))))...))))).....--.--. ( -36.40) >DroSim_CAF1 91582 110 - 1 GAGUACUGCGGCAACGUAGUUGGCUGCCACUUAAUGUGUUUUGCAUACUAAUUAGGUUGUUUGGCGGCUGCCGUCAC--------AUUUCAAGUGUUGAAACAGGUGACAGAAA--AAAA ....((((((....)))))).((((((((..((((.((.((........)).)).))))..))))))))...(((((--------.(((((.....)))))...))))).....--.... ( -34.80) >DroEre_CAF1 94587 112 - 1 GAGUACUGCGGCAACGUAGUUGGCUGCCACUUAAUGUGUUUUGCAUACUAAUUAGGUUGUUUGGCGGCUGCCGUCAC--------AUUUCAAGUGUUGAAACAGGUGACAGAAAUAAAAA ....((((((....)))))).((((((((..((((.((.((........)).)).))))..))))))))...(((((--------.(((((.....)))))...)))))........... ( -34.80) >DroYak_CAF1 93200 107 - 1 GAGUACUGCGGCAACGUAGUUGGCUGCCACUUAAUGUGUUUUGCAUACUAAUUAGGUUGUUUGGCGGCUGCCGUCAC--------AUUUCAAGUGUUGAAACAGGUGACAGAA-----AA ....((((((....)))))).((((((((..((((.((.((........)).)).))))..))))))))...(((((--------.(((((.....)))))...)))))....-----.. ( -34.80) >DroAna_CAF1 110226 116 - 1 GAGUACUGCGGCAACGUAGUUGGCUGCCACUUAAUGUGUUUUGCAUACUAAUUAGGUUGUUUGGCGGCUGCCGUCACAUUUUCACAUUUCAAGUGUUGGAACAGGUGACAGAAA--A--A ....((((((....)))))).((((((((..((((.((.((........)).)).))))..))))))))...(((((..((((((((.....))).)))))...))))).....--.--. ( -36.90) >consensus GAGUACUGCGGCAACGUAGUUGGCUGCCACUUAAUGUGUUUUGCAUACUAAUUAGGUUGUUUGGCGGCUGCCGUCAC________AUUUCAAGUGUUGAAACAGGUGACAGAAA__A__A ....((((((....)))))).((((((((((((((((((.....))))..))))))).....)))))))...(((((.........(((((.....)))))...)))))........... (-31.09 = -30.82 + -0.28)


| Location | 8,785,384 – 8,785,483 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.68 |
| Mean single sequence MFE | -24.73 |
| Consensus MFE | -21.39 |
| Energy contribution | -22.08 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
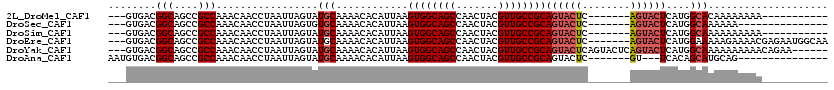
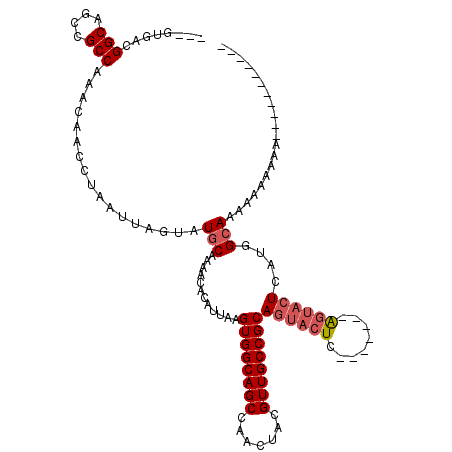
>2L_DroMel_CAF1 8785384 99 + 22407834 ---GUGACGGCAGCCGCCAAACAACCUAAUUAGUAUGCAAAACACAUUAAGUGGCAGCCAACUACGUUGCCGCAGUACUC-------AGUACUCAUGGCACAAAAAAAA----------- ---(((..(((....)))......((...(((((.((.....)).)))))((((((((.......))))))))(((((..-------.)))))...)))))........----------- ( -23.10) >DroSec_CAF1 88098 95 + 1 ---GUGACGGCAGCCGCCAAACAACCUAAUUAGUGUGCAAAACACAUUAAGUGGCAGCCAACUACGUUGCCGCAGUACUC-------AGUACUCAUGGCAAAAAA--------------- ---.....(((.(((((............((((((((.....))))))))))))).))).......((((((.(((((..-------.)))))..))))))....--------------- ( -28.70) >DroSim_CAF1 91615 99 + 1 ---GUGACGGCAGCCGCCAAACAACCUAAUUAGUAUGCAAAACACAUUAAGUGGCAGCCAACUACGUUGCCGCAGUACUC-------AGUACUCAUGGCAAAAAAAAAA----------- ---.....(((.(((((............(((((.((.....)).)))))))))).))).......((((((.(((((..-------.)))))..))))))........----------- ( -22.70) >DroEre_CAF1 94622 110 + 1 ---GUGACGGCAGCCGCCAAACAACCUAAUUAGUAUGCAAAACACAUUAAGUGGCAGCCAACUACGUUGCCGCAGUACUC-------AGUACUCAUGGAAAAAGAAAACGAGAAUGGCAA ---.....(((....))).................(((............((((((((.......))))))))(((((..-------.))))).......................))). ( -22.50) >DroYak_CAF1 93230 111 + 1 ---GUGACGGCAGCCGCCAAACAACCUAAUUAGUAUGCAAAACACAUUAAGUGGCAGCCAACUACGUUGCCGCAGUACUCAGUACUCAGUACUCAUGGCAAAAAAAAAACAGAA------ ---.....(((.(((((............(((((.((.....)).)))))))))).))).......((((((.((((((........))))))..)))))).............------ ( -25.00) >DroAna_CAF1 110262 95 + 1 AAUGUGACGGCAGCCGCCAAACAACCUAAUUAGUAUGCAAAACACAUUAAGUGGCAGCCAACUACGUUGCCGCAGUACUC-------GU---UCACAGCAUGCAG--------------- ..(((...(((....)))..))).........((((((............((((((((.......)))))))).((....-------..---..)).))))))..--------------- ( -26.40) >consensus ___GUGACGGCAGCCGCCAAACAACCUAAUUAGUAUGCAAAACACAUUAAGUGGCAGCCAACUACGUUGCCGCAGUACUC_______AGUACUCAUGGCAAAAAAAAAA___________ ........(((....))).................(((............((((((((.......))))))))((((((........))))))....))).................... (-21.39 = -22.08 + 0.69)
| Location | 8,785,384 – 8,785,483 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.68 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -25.91 |
| Energy contribution | -25.77 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.89 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
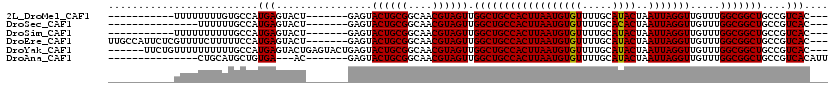
>2L_DroMel_CAF1 8785384 99 - 22407834 -----------UUUUUUUUGUGCCAUGAGUACU-------GAGUACUGCGGCAACGUAGUUGGCUGCCACUUAAUGUGUUUUGCAUACUAAUUAGGUUGUUUGGCGGCUGCCGUCAC--- -----------........((((.....))))(-------((..((((((....)))))).((((((((..((((.((.((........)).)).))))..))))))))....))).--- ( -28.00) >DroSec_CAF1 88098 95 - 1 ---------------UUUUUUGCCAUGAGUACU-------GAGUACUGCGGCAACGUAGUUGGCUGCCACUUAAUGUGUUUUGCACACUAAUUAGGUUGUUUGGCGGCUGCCGUCAC--- ---------------....(((((...(((((.-------..)))))..)))))....(..((((((((..(((((((.....))).((....))))))..))))))))..).....--- ( -27.60) >DroSim_CAF1 91615 99 - 1 -----------UUUUUUUUUUGCCAUGAGUACU-------GAGUACUGCGGCAACGUAGUUGGCUGCCACUUAAUGUGUUUUGCAUACUAAUUAGGUUGUUUGGCGGCUGCCGUCAC--- -----------..........((((..(((((.-------..)))))..((((((.(((((((.((((((.....)))....)))..))))))).))))))))))(((....)))..--- ( -26.40) >DroEre_CAF1 94622 110 - 1 UUGCCAUUCUCGUUUUCUUUUUCCAUGAGUACU-------GAGUACUGCGGCAACGUAGUUGGCUGCCACUUAAUGUGUUUUGCAUACUAAUUAGGUUGUUUGGCGGCUGCCGUCAC--- ..((((..(((((.(((.........))).)).-------))).((((((....)))))))))).((.(((((((((((.....))))..))))))).)).((((((...)))))).--- ( -29.40) >DroYak_CAF1 93230 111 - 1 ------UUCUGUUUUUUUUUUGCCAUGAGUACUGAGUACUGAGUACUGCGGCAACGUAGUUGGCUGCCACUUAAUGUGUUUUGCAUACUAAUUAGGUUGUUUGGCGGCUGCCGUCAC--- ------...............((((..((((((.(....).))))))..((((((.(((((((.((((((.....)))....)))..))))))).))))))))))(((....)))..--- ( -30.20) >DroAna_CAF1 110262 95 - 1 ---------------CUGCAUGCUGUGA---AC-------GAGUACUGCGGCAACGUAGUUGGCUGCCACUUAAUGUGUUUUGCAUACUAAUUAGGUUGUUUGGCGGCUGCCGUCACAUU ---------------........(((((---.(-------(.((((((((....)))))).((((((((..((((.((.((........)).)).))))..))))))))))))))))).. ( -32.80) >consensus ___________UUUUUUUUUUGCCAUGAGUACU_______GAGUACUGCGGCAACGUAGUUGGCUGCCACUUAAUGUGUUUUGCAUACUAAUUAGGUUGUUUGGCGGCUGCCGUCAC___ .........................(((................((((((....)))))).((((((((((((((((((.....))))..))))))).....)))))))....))).... (-25.91 = -25.77 + -0.14)
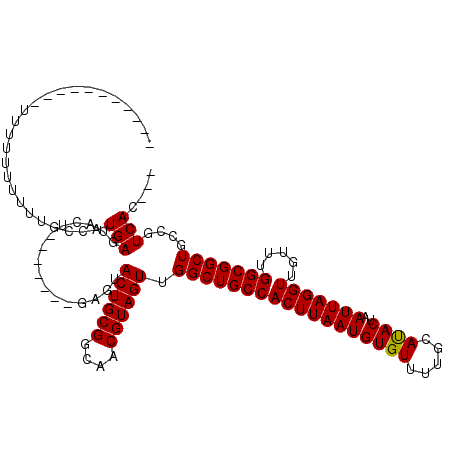
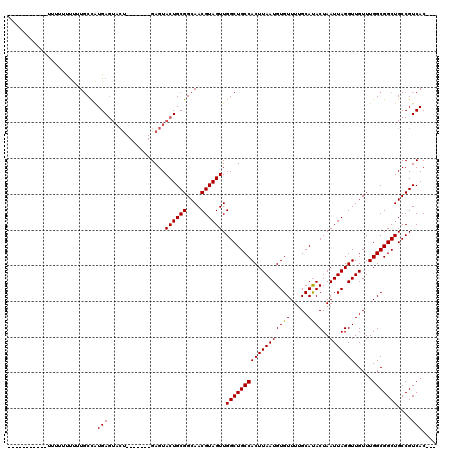
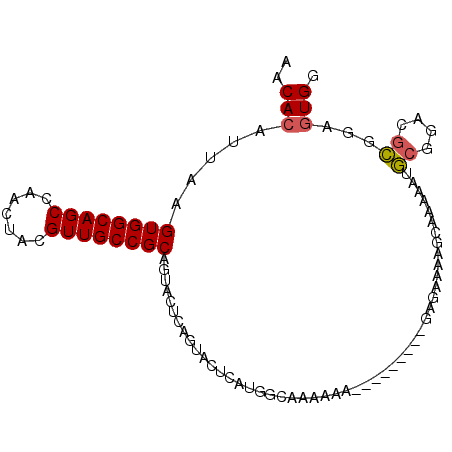
| Location | 8,785,421 – 8,785,514 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 75.04 |
| Mean single sequence MFE | -21.95 |
| Consensus MFE | -14.68 |
| Energy contribution | -14.80 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8785421 93 + 22407834 AACACAUUAAGUGGCAGCCAACUACGUUGCCGCAGUACUCAGUACUCAUGGCACAAAAAAAAA----AAAAAAACGAGAAAAGCGGACGCGGAUUGG ..(((.....)))....(((((..((((((((.(((((...)))))..))))...........----.......((.......))))))..).)))) ( -18.20) >DroSec_CAF1 88135 90 + 1 AACACAUUAAGUGGCAGCCAACUACGUUGCCGCAGUACUCAGUACUCAUGGCAAAAAA-------ACGAGAAAAGCAAAAAUGCGGACGCGGAGUGG ....((((..(((....((..((.((((((((.(((((...)))))..))))).....-------)))))....(((....))))).)))..)))). ( -22.00) >DroSim_CAF1 91652 97 + 1 AACACAUUAAGUGGCAGCCAACUACGUUGCCGCAGUACUCAGUACUCAUGGCAAAAAAAAAAAGUACGAGAAAAGCAAAAAUGCGGACGCGGAGUGG ..(((.....((((((((.......)))))))).((.(((.(((((................))))))))....(((....)))....))...))). ( -24.19) >DroAna_CAF1 110302 77 + 1 AACACAUUAAGUGGCAGCCAACUACGUUGCCGCAGUACUCGU---UCACAGCAUGCAG-----------------CUCGGAGUGGGCCAAUGAGUGG ..........((((((((.......))))))))..(((((((---(((((((.....)-----------------))....))))))....))))). ( -23.40) >consensus AACACAUUAAGUGGCAGCCAACUACGUUGCCGCAGUACUCAGUACUCAUGGCAAAAAA_________GAGAAAAGCAAAAAUGCGGACGCGGAGUGG ..(((.....((((((((.......)))))))).................................................((....))...))). (-14.68 = -14.80 + 0.12)

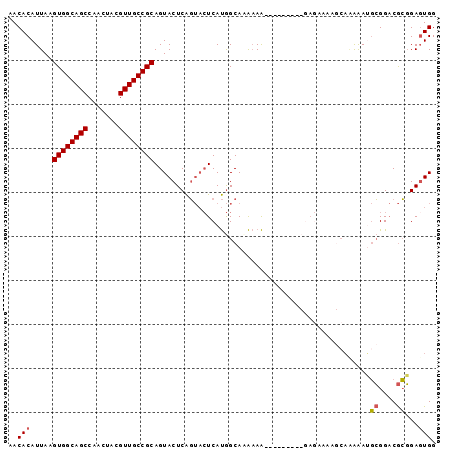
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:52 2006