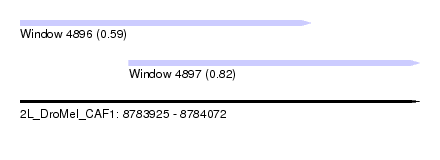
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,783,925 – 8,784,072 |
| Length | 147 |
| Max. P | 0.820496 |

| Location | 8,783,925 – 8,784,032 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.74 |
| Mean single sequence MFE | -35.26 |
| Consensus MFE | -24.77 |
| Energy contribution | -26.61 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8783925 107 + 22407834 CUUGUCGAAAUGUGUGCAAAUGGAGCACGCUGCAGUGCCGCAGGUCCUGGCUAAGGAUCUGCACACCUCCUCAUUUCGUUCUUGGCAGGGAACA-------------CUUCGCUCCACUC .....((((((((((((.......))))))....(((..(((((((((.....))))))))).)))......))))))....(((.((.((...-------------..)).)))))... ( -35.50) >DroSec_CAF1 86688 107 + 1 CUUGUCGAAAUGUGUGCAAAUGGAGCACGCUGCAGUGCCGCAGGUCCUGGCUAAGGAUCUGCACACCUUCUCACUUCGUUCUUGGCAGGGACCA-------------CUCCACUCCACUU ((((((((((((.(((.....((.((((......)))).(((((((((.....)))))))))...))....)))..)))).)))))))).....-------------............. ( -36.40) >DroSim_CAF1 90201 107 + 1 CUUGUCGAAAUGUGUGCAAAUGGAGCACGCUGCAGUGCCGCAGGUCCUGGCUAAGGAUCUGCACACCUUCUCAUUUCGUUCUUUGCAGGGACCA-------------CUCCGCUCCACUU ....................((((((...((((((((..(((((((((.....))))))))).))).................)))))(((...-------------.)))))))))... ( -34.92) >DroEre_CAF1 91842 107 + 1 CUUGUCGAAAUGUGUGCAAAUGGAGCACGCUGCAGUGCCGCAGGUCCUGGCUAAGGAUCUGCACACCUCCUUAUUCCCUUCUUGGCUGGCACCA-------------CUCCGCUCCACUC ...((.((...((((((.......))))))....((((((((((((((.....)))))))))..............((.....))..)))))..-------------.)).))....... ( -34.80) >DroYak_CAF1 91705 120 + 1 CUUGUCGAAAUGUGUGCAAAUGGAGCACGCUGCAGUGCCGCAGGUCCUGGCCAAGGAUCUGCACACCUCCUUACUUCGUUCUUGGCUGGGACCACCUAUCCCCUCAACUCCGCUCCACUU ....................((((((..((.((...)).)).(((((..(((((((((...................)))))))))..)))))..................))))))... ( -39.91) >DroAna_CAF1 108949 113 + 1 CCUGUCGAAAUGUGUGCAAAUGGAACACGCUGCAUUGCCCCUGGCACUGGCUAAGGAUCUCCAGCUCUGCCU-CUGCAUCCUUGA----GAAUAUCCUUCGGUU--GGUGCUCUCCACUC ....................((((....((......))....(((((..((((((((((((.((...(((..-..)))..)).))----))...))))).))).--.))))).))))... ( -30.00) >consensus CUUGUCGAAAUGUGUGCAAAUGGAGCACGCUGCAGUGCCGCAGGUCCUGGCUAAGGAUCUGCACACCUCCUCACUUCGUUCUUGGCAGGGACCA_____________CUCCGCUCCACUC ....................((((((..((.((...)).)).(((((..(((((((((...................)))))))))..)))))..................))))))... (-24.77 = -26.61 + 1.84)

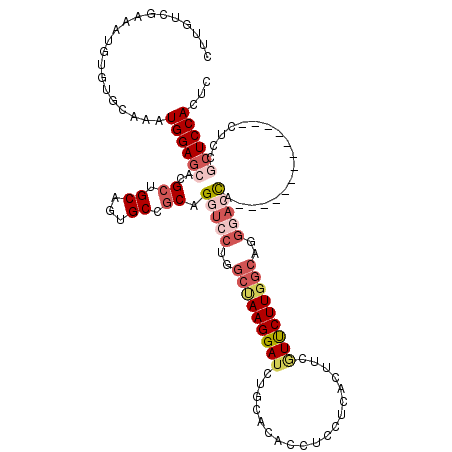
| Location | 8,783,965 – 8,784,072 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.47 |
| Mean single sequence MFE | -30.19 |
| Consensus MFE | -20.82 |
| Energy contribution | -22.41 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8783965 107 + 22407834 CAGGUCCUGGCUAAGGAUCUGCACACCUCCUCAUUUCGUUCUUGGCAGGGAACA-------------CUUCGCUCCACUCCGGCACUGCAGUAAAAUAUUGUUUCGUCCUGCUCGAGUAU ((((((((.....))))))))...................(((((((((((.(.-------------....).))).....(((...((((((...))))))...))))))).))))... ( -27.70) >DroSec_CAF1 86728 107 + 1 CAGGUCCUGGCUAAGGAUCUGCACACCUUCUCACUUCGUUCUUGGCAGGGACCA-------------CUCCACUCCACUUCGGCACUGCAGUAAAAUAUUGUUUCGUCCUGCUCGAGUAU ..((((((.(((((((((...................))))))))).)))))).-------------....((((......(((...((((((...))))))...)))......)))).. ( -28.91) >DroSim_CAF1 90241 107 + 1 CAGGUCCUGGCUAAGGAUCUGCACACCUUCUCAUUUCGUUCUUUGCAGGGACCA-------------CUCCGCUCCACUUCGGCACUGCAGUAAAAUAUUGUUUCGUCCUGCUCGAGUAU ((((((((.....)))))))).............((((......(((((((.(.-------------....).))).....(((...((((((...))))))...))))))).))))... ( -25.10) >DroEre_CAF1 91882 107 + 1 CAGGUCCUGGCUAAGGAUCUGCACACCUCCUUAUUCCCUUCUUGGCUGGCACCA-------------CUCCGCUCCACUCCGGCACUGCAGUAAAAUAUUGUUUCGUGCCGCUCGAGUAU ..(((.(..((((((((.....................))))))))..).))).-------------....((((.....((((((.((((((...))))))...))))))...)))).. ( -32.40) >DroYak_CAF1 91745 120 + 1 CAGGUCCUGGCCAAGGAUCUGCACACCUCCUUACUUCGUUCUUGGCUGGGACCACCUAUCCCCUCAACUCCGCUCCACUUUGGCACUGCAGUAAAAUAUUGUUUCGUUCCGCUCGAGUAU ..(((((..(((((((((...................)))))))))..)))))..................((((.....(((.((.((((((...))))))...)).)))...)))).. ( -34.51) >DroAna_CAF1 108989 111 + 1 CUGGCACUGGCUAAGGAUCUCCAGCUCUGCCU-CUGCAUCCUUGA----GAAUAUCCUUCGGUU--GGUGCUCUCCACUCCACCAAUGCAGUAGGA-AUUGUCUCGC-CCGUACGAGUAU ..(((((..((((((((((((.((...(((..-..)))..)).))----))...))))).))).--.))))).((((((.((....)).))).)))-.....((((.-.....))))... ( -32.50) >consensus CAGGUCCUGGCUAAGGAUCUGCACACCUCCUCACUUCGUUCUUGGCAGGGACCA_____________CUCCGCUCCACUCCGGCACUGCAGUAAAAUAUUGUUUCGUCCCGCUCGAGUAU ..(((((..(((((((((...................)))))))))..))))).......................((((.(((((.(((((.....)))))...))...))).)))).. (-20.82 = -22.41 + 1.59)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:48 2006