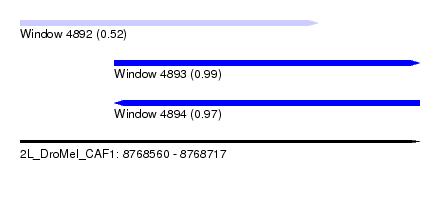
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,768,560 – 8,768,717 |
| Length | 157 |
| Max. P | 0.989278 |

| Location | 8,768,560 – 8,768,677 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.26 |
| Mean single sequence MFE | -25.72 |
| Consensus MFE | -20.00 |
| Energy contribution | -19.52 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8768560 117 + 22407834 AAGAUCAAAGAUCUAUUGUUUUUCUCACCUCAUA---ACAAACAGCCUCAGAUUGGACAAUAUAAGAGGCAAACAAGGCCAACAAAAUGGCCAAGUUCUUUACAGAACUCAUCUUGCAGA .(((((...))))).(((((.............)---))))...(((((..((........))..)))))......(((((......))))).((((((....))))))........... ( -24.92) >DroSec_CAF1 72805 117 + 1 ---UUCAAGGAUCUCAUGUUUUUCUCACCUCAUAGCCACAAACAGCCUCGGAUUGCACAAUAUAGGAGGCAAACAAGGCCAACAAAAUGGCCAAGUUCUUCACAGAACUCAUCUUGCAGA ---..(((((..................................(((((..((........))..)))))......(((((......))))).((((((....))))))..))))).... ( -24.40) >DroSim_CAF1 75436 117 + 1 ---UUCAAGGAUCUCAUGUUUUUCUCACCUCAUAACCACAAACAGCCUCGGAUUGCACAAUAUAGGAGGCAAACAAGGCCAACAAAAUGGCCAAGUUCUUCACAGAACUCAUCUUGCAGA ---..(((((..................................(((((..((........))..)))))......(((((......))))).((((((....))))))..))))).... ( -24.40) >DroEre_CAF1 76875 117 + 1 ---UCCAAAGAUCUCUUGUUCUGCUCACCUCAUAGCCACAAACAGCCUCGGAUUGCGUGAUAUAAGAGGCAAACAAGGCCAACAAAAUGGCAAAAGUCUUCAAGGAACUCAUCUUGCUGA ---(((.(((((..((((((..(((........)))........(((((..((........))..))))).))))))((((......))))....)))))...))).............. ( -25.10) >DroYak_CAF1 76135 116 + 1 ---UU-AAAGGACUCUUGUUUUGCUCACCUCGUAACCACACACAGCCUCUGAUGGCGUGACAUGAGAGGCGAUCAAGGCCAAGAAAAUGGCCAAGAUAUUUACAGAACCCAUUUUGCAGA ---..-...........((((((.....................((((((.(((......))).)))))).(((..(((((......)))))..))).....))))))............ ( -29.80) >consensus ___UUCAAAGAUCUCUUGUUUUUCUCACCUCAUAACCACAAACAGCCUCGGAUUGCACAAUAUAAGAGGCAAACAAGGCCAACAAAAUGGCCAAGUUCUUCACAGAACUCAUCUUGCAGA .......(((((.....(((((......................(((((..((........))..)))))......(((((......)))))...........)))))..)))))..... (-20.00 = -19.52 + -0.48)

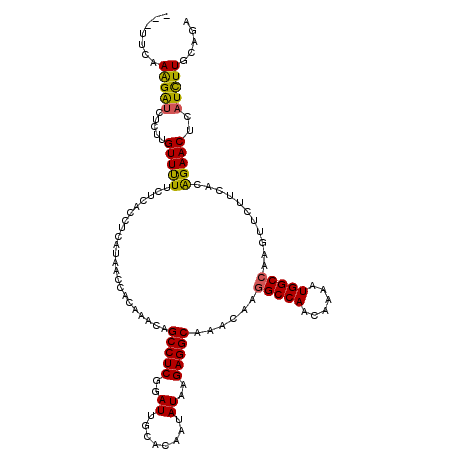
| Location | 8,768,597 – 8,768,717 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.11 |
| Mean single sequence MFE | -31.85 |
| Consensus MFE | -14.30 |
| Energy contribution | -16.08 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.45 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8768597 120 + 22407834 AACAGCCUCAGAUUGGACAAUAUAAGAGGCAAACAAGGCCAACAAAAUGGCCAAGUUCUUUACAGAACUCAUCUUGCAGAGUUCCCUAACCAGCAACUGCACAACAAGAACUGGCUAUAC ....(((((..((........))..)))))................(((((((.((((((....((((((........))))))........((....)).....))))))))))))).. ( -32.40) >DroSec_CAF1 72842 120 + 1 AACAGCCUCGGAUUGCACAAUAUAGGAGGCAAACAAGGCCAACAAAAUGGCCAAGUUCUUCACAGAACUCAUCUUGCAGAGUUCACUAAGCAGCAACUACACAACAAGAACUGGCUAUAC ....((((.(..((((.(......)...)))).).)))).......(((((((.((((((....((((((........))))))....((......)).......))))))))))))).. ( -30.60) >DroSim_CAF1 75473 120 + 1 AACAGCCUCGGAUUGCACAAUAUAGGAGGCAAACAAGGCCAACAAAAUGGCCAAGUUCUUCACAGAACUCAUCUUGCAGAGUUCACUAAGCAGCAACUGCACAACAAGAACUGGCUAUAC ....((((.(..((((.(......)...)))).).)))).......(((((((.((((((....((((((........)))))).....((((...)))).....))))))))))))).. ( -36.10) >DroEre_CAF1 76912 120 + 1 AACAGCCUCGGAUUGCGUGAUAUAAGAGGCAAACAAGGCCAACAAAAUGGCAAAAGUCUUCAAGGAACUCAUCUUGCUGAGUGCCCUAACCAACGACUAAAAGCCAAGAACUGGCCUUAC ....(((((..((........))..)))))....(((((((......((((...((((....(((.(((((......)))))..))).......))))....)))).....))))))).. ( -35.50) >DroYak_CAF1 76171 120 + 1 CACAGCCUCUGAUGGCGUGACAUGAGAGGCGAUCAAGGCCAAGAAAAUGGCCAAGAUAUUUACAGAACCCAUUUUGCAGAGUUCCCUAACCAACGACUACAAACCAAGAACUGGCUUUAC ....((((((.(((......))).)))))).(((..(((((......)))))..)))..................((..(((((.......................))))).))..... ( -29.80) >DroAna_CAF1 74030 120 + 1 AACAGAUUCACUUUGGCUGAUGUAAAAGCCACAAAGGACUAGCAGGAUACCGAAGGCUUUCACCUUCGCCAUUGUUCGGAGUUCUCGAUCUAUCGUUUUCACGGAAAAUACUGACUUCAU ..(((..((....(((((........)))))...((((((....(((((.((((((......))))))....)))))..)))))).))......((((((...)))))).)))....... ( -26.70) >consensus AACAGCCUCGGAUUGCACAAUAUAAGAGGCAAACAAGGCCAACAAAAUGGCCAAGUUCUUCACAGAACUCAUCUUGCAGAGUUCCCUAACCAGCAACUACACAACAAGAACUGGCUAUAC ....(((((..((........))..)))))......(((((......)))))............((((((........)))))).................................... (-14.30 = -16.08 + 1.78)


| Location | 8,768,597 – 8,768,717 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.11 |
| Mean single sequence MFE | -37.78 |
| Consensus MFE | -20.28 |
| Energy contribution | -21.04 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8768597 120 - 22407834 GUAUAGCCAGUUCUUGUUGUGCAGUUGCUGGUUAGGGAACUCUGCAAGAUGAGUUCUGUAAAGAACUUGGCCAUUUUGUUGGCCUUGUUUGCCUCUUAUAUUGUCCAAUCUGAGGCUGUU ...((((((((..(((.....)))..)))))))).(((((((........))))))).....((((..(((((......)))))..))))(((((..((........))..))))).... ( -42.40) >DroSec_CAF1 72842 120 - 1 GUAUAGCCAGUUCUUGUUGUGUAGUUGCUGCUUAGUGAACUCUGCAAGAUGAGUUCUGUGAAGAACUUGGCCAUUUUGUUGGCCUUGUUUGCCUCCUAUAUUGUGCAAUCCGAGGCUGUU ..((((((.....((((.((((((..(((((..((....))..)))....(((((((....)))))))(((((......)))))......))...))))))...)))).....)))))). ( -36.20) >DroSim_CAF1 75473 120 - 1 GUAUAGCCAGUUCUUGUUGUGCAGUUGCUGCUUAGUGAACUCUGCAAGAUGAGUUCUGUGAAGAACUUGGCCAUUUUGUUGGCCUUGUUUGCCUCCUAUAUUGUGCAAUCCGAGGCUGUU ..((((((...((..((((..((((....((.....((((((........))))))......((((..(((((......)))))..)))))).......))))..))))..)))))))). ( -37.90) >DroEre_CAF1 76912 120 - 1 GUAAGGCCAGUUCUUGGCUUUUAGUCGUUGGUUAGGGCACUCAGCAAGAUGAGUUCCUUGAAGACUUUUGCCAUUUUGUUGGCCUUGUUUGCCUCUUAUAUCACGCAAUCCGAGGCUGUU .(((((((((....((((....((((.....((((((.(((((......))))).)))))).))))...)))).....)))))))))...(((((................))))).... ( -42.89) >DroYak_CAF1 76171 120 - 1 GUAAAGCCAGUUCUUGGUUUGUAGUCGUUGGUUAGGGAACUCUGCAAAAUGGGUUCUGUAAAUAUCUUGGCCAUUUUCUUGGCCUUGAUCGCCUCUCAUGUCACGCCAUCAGAGGCUGUG ...(((((((...)))))))...........(((.(((((((........))))))).)))..(((..(((((......)))))..))).((((((.(((......))).)))))).... ( -38.40) >DroAna_CAF1 74030 120 - 1 AUGAAGUCAGUAUUUUCCGUGAAAACGAUAGAUCGAGAACUCCGAACAAUGGCGAAGGUGAAAGCCUUCGGUAUCCUGCUAGUCCUUUGUGGCUUUUACAUCAGCCAAAGUGAAUCUGUU .................(((....)))((((((((.(....))))((..((((((((((....)))))).(((....((((........))))...)))....))))..))...))))). ( -28.90) >consensus GUAAAGCCAGUUCUUGUUGUGUAGUCGCUGGUUAGGGAACUCUGCAAGAUGAGUUCUGUGAAGAACUUGGCCAUUUUGUUGGCCUUGUUUGCCUCUUAUAUCAUGCAAUCCGAGGCUGUU ....(((((((..(((.....)))..)))))))...((((((........))))))......((((..(((((......)))))..))))(((((................))))).... (-20.28 = -21.04 + 0.76)


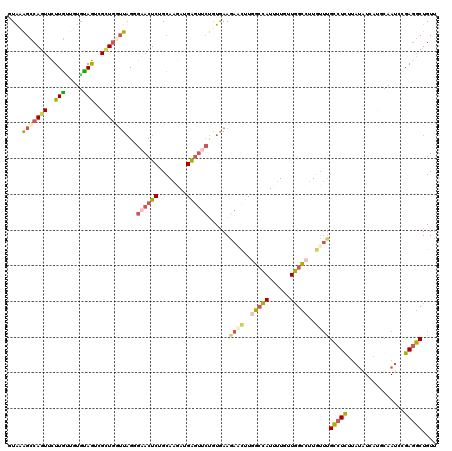
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:45 2006