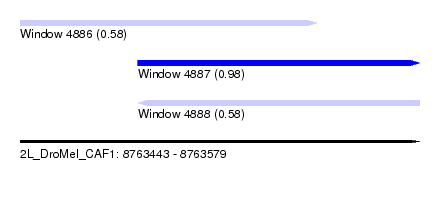
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,763,443 – 8,763,579 |
| Length | 136 |
| Max. P | 0.976934 |

| Location | 8,763,443 – 8,763,544 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.93 |
| Mean single sequence MFE | -29.66 |
| Consensus MFE | -24.66 |
| Energy contribution | -24.97 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
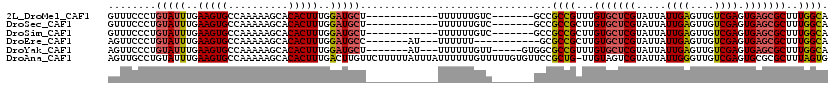
>2L_DroMel_CAF1 8763443 101 + 22407834 GUUUCCCUGUAUUUGAAGUGCCAAAAAGCACACUUUGGAUGCU------------UUUUUUGUC-------GCCGCCGUUUGUGCUCGUAUUAUUGAGUUGUCGAGUGAGCGCUUUGGCA ........(((((..(((((..........)))))..))))).------------.........-------...((((...(((((((.....((((....)))).)))))))..)))). ( -29.00) >DroSec_CAF1 67720 101 + 1 GUUUCCCUGUAUUUGAAGUGCCAAAAAGCACACUUUGGAUGCU------------UUUUUUGUC-------GCCGCCGCUUGUGCUCGUAUUAUUGAGUUGUCGAGUGAGCGCUUUGGCA ........(((((..(((((..........)))))..))))).------------.....((((-------(.(((((((((.(((((......)))))...)))))).)))...))))) ( -31.80) >DroSim_CAF1 70335 101 + 1 GUUUCCCUGUAUUUGAAGUGCCAAAAAGCACACUUUGGAUGCU------------UUUUUUGUC-------GCCGCCGCUUGUGCUCGUAUUAUUGAGUUGUCGAGUGAGCGCUUUGGCA ........(((((..(((((..........)))))..))))).------------.....((((-------(.(((((((((.(((((......)))))...)))))).)))...))))) ( -31.80) >DroEre_CAF1 71676 99 + 1 AGUUCCCUGUAUUUGAAGUGCCAAAAAGCACACUUUGGAUGCC-------AU---UUUUUU-----------GCGCCGCUUGUGCUCGUAUUAUUGAGUUGUCGAGUGAGCGCUUUGGCA ..........(((..(((((..........)))))..)))(((-------(.---......-----------((((((((((.(((((......)))))...)))))).))))..)))). ( -33.40) >DroYak_CAF1 70700 105 + 1 AGUUCCCUGUAUUUGAAGUGCCAAAAAGCACACUUUGGAUGCU-------AU---UUUUUUGUU-----GUGGCGCCGUUUGUGCUCGUAUUAUUGAGUUGUCGAGUGAGCGCUUUGGCA ........(((((..(((((..........)))))..))))).-------..---.....((((-----(.(((((((((((.(((((......)))))...)))))).))))).))))) ( -31.00) >DroAna_CAF1 69252 119 + 1 AGUUGCCUGUAUUUGAAGUGCCAAAAAGCACACUUUGACUUGUUCUUUUUAUUUAUUUUUUGUUUUUGUGUUCCGCUG-UUGUAGUCGUAUUAUUGGGUUGUCGAGUGCGCGCUUUAGUG ............(((((((((.....(((((((...(((......................)))...))))...))).-..(((.(((.(((....)))...))).)))))))))))).. ( -20.95) >consensus AGUUCCCUGUAUUUGAAGUGCCAAAAAGCACACUUUGGAUGCU____________UUUUUUGUC_______GCCGCCGCUUGUGCUCGUAUUAUUGAGUUGUCGAGUGAGCGCUUUGGCA ........(((((..(((((..........)))))..)))))................................((((...(((((((.....((((....)))).)))))))..)))). (-24.66 = -24.97 + 0.31)

| Location | 8,763,483 – 8,763,579 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 93.26 |
| Mean single sequence MFE | -31.06 |
| Consensus MFE | -25.02 |
| Energy contribution | -25.26 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8763483 96 + 22407834 GCU--UUUUUUGUC--GCCGCCGUUUGUGCUCGUAUUAUUGAGUUGUCGAGUGAGCGCUUUGGCAAGGAUUUAUGCUCGAUGGCG---AUAUUUGUUUAGCCG (((--.....((((--(((((((...(((((((.....((((....)))).)))))))..))))...((.......))...))))---))).......))).. ( -29.20) >DroSec_CAF1 67760 96 + 1 GCU--UUUUUUGUC--GCCGCCGCUUGUGCUCGUAUUAUUGAGUUGUCGAGUGAGCGCUUUGGCAAGGAUUUAUGCUCGAUGGCG---AUAUUUGUUUAGCCG (((--.....((((--(((((((((((.(((((......)))))...)))))).))...((((((........))).))).))))---))).......))).. ( -32.00) >DroSim_CAF1 70375 96 + 1 GCU--UUUUUUGUC--GCCGCCGCUUGUGCUCGUAUUAUUGAGUUGUCGAGUGAGCGCUUUGGCAAGGAUUUAUGCUCGAUGGCG---AUAUUUGUUUAGCCG (((--.....((((--(((((((((((.(((((......)))))...)))))).))...((((((........))).))).))))---))).......))).. ( -32.00) >DroEre_CAF1 71716 94 + 1 GCCAUUUUUUU------GCGCCGCUUGUGCUCGUAUUAUUGAGUUGUCGAGUGAGCGCUUUGGCAAGGAUUUAUGCUCGAUGGCG---AUAUUUGUUUAGCCG ((((((.....------((((((((((.(((((......)))))...)))))).))))...((((........)))).)))))).---............... ( -31.00) >DroYak_CAF1 70740 103 + 1 GCUAUUUUUUUGUUGUGGCGCCGUUUGUGCUCGUAUUAUUGAGUUGUCGAGUGAGCGCUUUGGCAAGGAUUUAUGCUCGAUGGCGAUGAUAUUUGUUUAGCCG ((((..(((((((((.(((((((((((.(((((......)))))...)))))).))))).)))))))))..)).)).....(((...(((....)))..))). ( -31.10) >consensus GCU__UUUUUUGUC__GCCGCCGCUUGUGCUCGUAUUAUUGAGUUGUCGAGUGAGCGCUUUGGCAAGGAUUUAUGCUCGAUGGCG___AUAUUUGUUUAGCCG ((....((((((((....(((((((((.(((((......)))))...)))))).)))....)))))))).....)).....(((...............))). (-25.02 = -25.26 + 0.24)

| Location | 8,763,483 – 8,763,579 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 93.26 |
| Mean single sequence MFE | -17.56 |
| Consensus MFE | -14.41 |
| Energy contribution | -14.41 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8763483 96 - 22407834 CGGCUAAACAAAUAU---CGCCAUCGAGCAUAAAUCCUUGCCAAAGCGCUCACUCGACAACUCAAUAAUACGAGCACAAACGGCGGC--GACAAAAAA--AGC ..............(---((((..((.(((........)))....(.((((....................)))).)...))..)))--)).......--... ( -18.25) >DroSec_CAF1 67760 96 - 1 CGGCUAAACAAAUAU---CGCCAUCGAGCAUAAAUCCUUGCCAAAGCGCUCACUCGACAACUCAAUAAUACGAGCACAAGCGGCGGC--GACAAAAAA--AGC ..............(---((((...(.(((........))))....((((..((((..............))))....))))..)))--)).......--... ( -18.54) >DroSim_CAF1 70375 96 - 1 CGGCUAAACAAAUAU---CGCCAUCGAGCAUAAAUCCUUGCCAAAGCGCUCACUCGACAACUCAAUAAUACGAGCACAAGCGGCGGC--GACAAAAAA--AGC ..............(---((((...(.(((........))))....((((..((((..............))))....))))..)))--)).......--... ( -18.54) >DroEre_CAF1 71716 94 - 1 CGGCUAAACAAAUAU---CGCCAUCGAGCAUAAAUCCUUGCCAAAGCGCUCACUCGACAACUCAAUAAUACGAGCACAAGCGGCGC------AAAAAAAUGGC ...............---.(((((.(.(((........))))...((((.(.((.(....(((........)))..).)).)))))------......))))) ( -17.40) >DroYak_CAF1 70740 103 - 1 CGGCUAAACAAAUAUCAUCGCCAUCGAGCAUAAAUCCUUGCCAAAGCGCUCACUCGACAACUCAAUAAUACGAGCACAAACGGCGCCACAACAAAAAAAUAGC .(((((......)).....(((...(.(((........))))...(.((((....................)))).)....))))))................ ( -15.05) >consensus CGGCUAAACAAAUAU___CGCCAUCGAGCAUAAAUCCUUGCCAAAGCGCUCACUCGACAACUCAAUAAUACGAGCACAAGCGGCGGC__GACAAAAAA__AGC ..................((((...(.(((........))))...(.((((....................)))).)....)))).................. (-14.41 = -14.41 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:39 2006