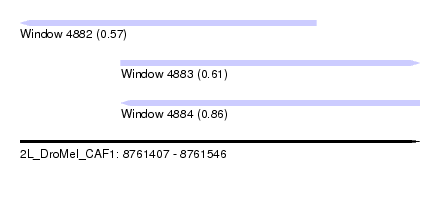
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,761,407 – 8,761,546 |
| Length | 139 |
| Max. P | 0.861339 |

| Location | 8,761,407 – 8,761,510 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.90 |
| Mean single sequence MFE | -31.27 |
| Consensus MFE | -17.19 |
| Energy contribution | -16.28 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
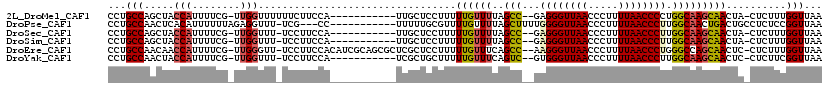
>2L_DroMel_CAF1 8761407 103 - 22407834 --------UUGCUCCUUUUUGUU-UUAGCC--GAGGGUUAACCCUUUUAACCCCUGGCAAGCAACUA-CUCUUUGGUUAACCACGGACAGCGCUGG-AUGACAUUUUUUCC----AGGAC --------..(((.....(((((-(..(((--(.(((((((.....))))))).))))))))))...-.(((.(((....))).))).))).((((-(..........)))----))... ( -30.50) >DroPse_CAF1 86224 110 - 1 --------UUUUUGCGUUUUGUU-UUAGCUUUUGGGGUUAACCCUUUUAACCCUUGGCAACUGACUGCCUCUCCGGUUAACCACGGACAGCGUUAGCCUGACAUUU-UUCCAGGCAGGAC --------.......((((((..-...(((...((((((((.....)))))))).)))..(((((.((...((((........))))..)))))))((((......-...)))))))))) ( -30.70) >DroSim_CAF1 68284 102 - 1 --------UUGCUCCUUUUUGUU-UUAGCC--GAGGGUUAACCCUUUUAACCCUUGGCAAGCAACUA-CUCUUUGGUUAACCACGGACAGCGCUGG-AUGACAUUU-UUCC----AGGAC --------..(((.....(((((-(..(((--(((((((((.....))))))))))))))))))...-.(((.(((....))).))).))).((((-(........-.)))----))... ( -35.10) >DroEre_CAF1 69522 110 - 1 CGCAGCGCUCGCUCCUUUUUGUU-UCAGCC--AAGGGUUAACCCUUUUAACCCUGGGCCAGCAACUC-CUCUUUGGUUAACCACGGACAGCGCUGG-AUGACAUUU-UUCC----AGGAC ..(((((((.(.(((...(((((-...(((--.((((((((.....)))))))).))).)))))...-.....(((....))).))))))))))).-.........-....----..... ( -35.30) >DroYak_CAF1 68553 102 - 1 --------UCGCUGCUUUUUGUU-UCAGUC--GUGGGUUAACCCUUUUAACCCUUGGCAAGCAACUC-CUCUUCGGUUAACCACGGACAGCGCUGG-AUGACAUUU-UUCC----AGCAC --------..((((((..(((((-(..(((--(.(((((((.....))))))).))))))))))...-......((....))..)).))))(((((-(........-.)))----))).. ( -32.90) >DroAna_CAF1 67287 100 - 1 --------U--AUCCUUUCUGUUUUUAGCA--AGGGGUUAACUCUUUUAACCCUUGGCAAGCAACUA-CUCUUCGGUUAACCACGGACAACGUUAG-AUGACAUU--UUCC----AGGAC --------.--.((((...(((((...((.--(((((((((.....))))))))).)))))))....-......((....))..(((.((.(((..-..))).))--.)))----)))). ( -23.10) >consensus ________UUGCUCCUUUUUGUU_UUAGCC__GAGGGUUAACCCUUUUAACCCUUGGCAAGCAACUA_CUCUUCGGUUAACCACGGACAGCGCUGG_AUGACAUUU_UUCC____AGGAC ............((((...((((.(((((.....(((((((.....)))))))...............((.((((........)))).)).)))))...))))............)))). (-17.19 = -16.28 + -0.91)

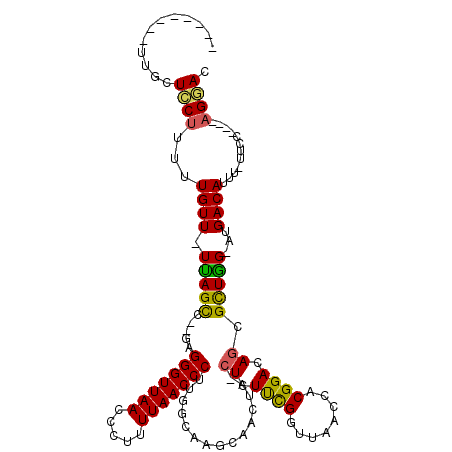
| Location | 8,761,442 – 8,761,546 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.77 |
| Mean single sequence MFE | -27.02 |
| Consensus MFE | -16.35 |
| Energy contribution | -17.68 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8761442 104 + 22407834 UUAACCAAAGAG-UAGUUGCUUGCCAGGGGUUAAAAGGGUUAACCCUC--GGCUAAAACAAAAAGGAGCAA-----------UGGAAGAAAAAACCAA-CGAAAAUGGUAGCUGGCAGG ....(((.....-..((((((((((..(((((((.....)))))))..--))).....(.....)))))))-----------)..........((((.-......))))...))).... ( -25.60) >DroPse_CAF1 86263 104 + 1 UUAACCGGAGAGGCAGUCAGUUGCCAAGGGUUAAAAGGGUUAACCCCAAAAGCUAAAACAAAACGCAAAAA-----------GG---CGA-AAACCUCUAAAAAAUGUGAGUUGGCAGG ....((.....(((((....)))))..(((((((.....))))))).....(((((.......(((.....-----------.)---)).-....(((..........)))))))).)) ( -23.90) >DroSec_CAF1 65683 103 + 1 UUAACCAAAGAG-UAGUUGCUUGCCAAGGGUUAAAAGGGUUAACCCUC--GGCUAAAACAAAAAGGAGCAA-----------UGGAAGGA-AAACCAA-CGAAAAUGGUAGCUGGCAGG ....((......-..((((((((((.((((((((.....)))))))).--))).....(.....)))))))-----------).(..((.-..((((.-......))))..))..).)) ( -26.20) >DroSim_CAF1 68318 103 + 1 UUAACCAAAGAG-UAGUUGCUUGCCAAGGGUUAAAAGGGUUAACCCUC--GGCUAAAACAAAAAGGAGCAA-----------UGGAAGGA-AAACCAA-CGAAAAUGGUAGCUGGCAGG ....((......-..((((((((((.((((((((.....)))))))).--))).....(.....)))))))-----------).(..((.-..((((.-......))))..))..).)) ( -26.20) >DroEre_CAF1 69556 114 + 1 UUAACCAAAGAG-GAGUUGCUGGCCCAGGGUUAAAAGGGUUAACCCUU--GGCUGAAACAAAAAGGAGCGAGCGCUGCGAUGUGGAAGGA-AACCCAA-CGAAAAUGGUUGUUGGCAGG ....(((....(-.(((.(((.((((.(((((((.....)))))))((--(.......)))...)).)).)))))).)....)))..(..-..)((((-(((......))))))).... ( -34.90) >DroYak_CAF1 68587 103 + 1 UUAACCGAAGAG-GAGUUGCUUGCCAAGGGUUAAAAGGGUUAACCCAC--GACUGAAACAAAAAGCAGCGA-----------UGGAAGGA-AAACCAA-CGAAAAUGGUAGUUGGCAGG ....((.....)-).....(((((((((((((((.....))))))).(--(.(((..........))))).-----------........-..((((.-......))))..)))))))) ( -25.30) >consensus UUAACCAAAGAG_UAGUUGCUUGCCAAGGGUUAAAAGGGUUAACCCUC__GGCUAAAACAAAAAGGAGCAA___________UGGAAGGA_AAACCAA_CGAAAAUGGUAGCUGGCAGG ...................(((((((((((((((.....)))))))...............................................((((........))))..)))))))) (-16.35 = -17.68 + 1.33)

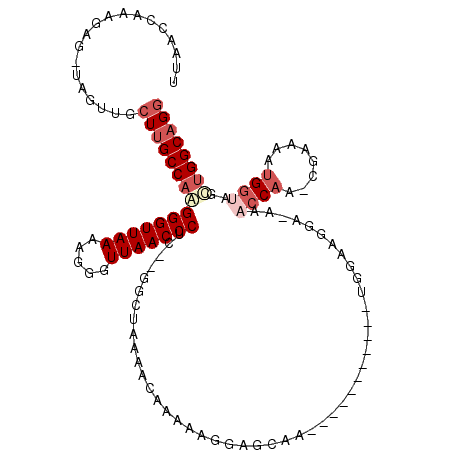
| Location | 8,761,442 – 8,761,546 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.77 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -16.89 |
| Energy contribution | -17.25 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8761442 104 - 22407834 CCUGCCAGCUACCAUUUUCG-UUGGUUUUUUCUUCCA-----------UUGCUCCUUUUUGUUUUAGCC--GAGGGUUAACCCUUUUAACCCCUGGCAAGCAACUA-CUCUUUGGUUAA ...((((((..........)-)))))........(((-----------..(.......((((((..(((--(.(((((((.....))))))).))))))))))...-..)..))).... ( -24.50) >DroPse_CAF1 86263 104 - 1 CCUGCCAACUCACAUUUUUUAGAGGUUU-UCG---CC-----------UUUUUGCGUUUUGUUUUAGCUUUUGGGGUUAACCCUUUUAACCCUUGGCAACUGACUGCCUCUCCGGUUAA ..((((((...........((((((((.-.((---(.-----------.....))).........))))))))(((((((.....)))))))))))))..((((((......)))))). ( -24.60) >DroSec_CAF1 65683 103 - 1 CCUGCCAGCUACCAUUUUCG-UUGGUUU-UCCUUCCA-----------UUGCUCCUUUUUGUUUUAGCC--GAGGGUUAACCCUUUUAACCCUUGGCAAGCAACUA-CUCUUUGGUUAA ...((((((..........)-)))))..-.....(((-----------..(.......((((((..(((--(((((((((.....))))))))))))))))))...-..)..))).... ( -29.00) >DroSim_CAF1 68318 103 - 1 CCUGCCAGCUACCAUUUUCG-UUGGUUU-UCCUUCCA-----------UUGCUCCUUUUUGUUUUAGCC--GAGGGUUAACCCUUUUAACCCUUGGCAAGCAACUA-CUCUUUGGUUAA ...((((((..........)-)))))..-.....(((-----------..(.......((((((..(((--(((((((((.....))))))))))))))))))...-..)..))).... ( -29.00) >DroEre_CAF1 69556 114 - 1 CCUGCCAACAACCAUUUUCG-UUGGGUU-UCCUUCCACAUCGCAGCGCUCGCUCCUUUUUGUUUCAGCC--AAGGGUUAACCCUUUUAACCCUGGGCCAGCAACUC-CUCUUUGGUUAA .........(((((.....(-(.(((..-....)))))...(((((....))).............(((--.((((((((.....)))))))).)))..)).....-.....))))).. ( -27.40) >DroYak_CAF1 68587 103 - 1 CCUGCCAACUACCAUUUUCG-UUGGUUU-UCCUUCCA-----------UCGCUGCUUUUUGUUUCAGUC--GUGGGUUAACCCUUUUAACCCUUGGCAAGCAACUC-CUCUUCGGUUAA ...((((((..........)-)))))..-.......(-----------(((..(....((((((..(((--(.(((((((.....))))))).))))))))))...-..)..))))... ( -21.50) >consensus CCUGCCAACUACCAUUUUCG_UUGGUUU_UCCUUCCA___________UUGCUCCUUUUUGUUUUAGCC__GAGGGUUAACCCUUUUAACCCUUGGCAAGCAACUA_CUCUUUGGUUAA ...(((.....(((........))).................................((((((..(((...((((((((.....)))))))).)))))))))..........)))... (-16.89 = -17.25 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:35 2006