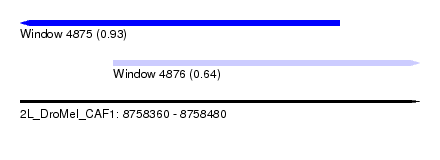
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,758,360 – 8,758,480 |
| Length | 120 |
| Max. P | 0.927533 |

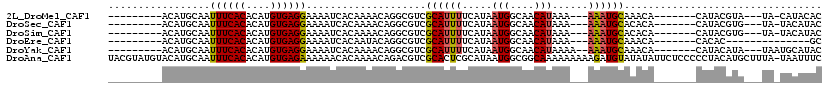
| Location | 8,758,360 – 8,758,456 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.89 |
| Mean single sequence MFE | -18.73 |
| Consensus MFE | -12.10 |
| Energy contribution | -12.18 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8758360 96 - 22407834 ---------ACAUGCAAUUUCACACAUGUGAGGAAAAUCACAAAACAGGCGUCGCAUUUUCAUAAUGGCAACAUAAA---AAAUGCAAACA-------CAUACGUA---UA-CAUACAC ---------..((((...........(((((......)))))...........(((((((....(((....)))..)---)))))).....-------.....)))---).-....... ( -17.00) >DroSec_CAF1 62675 96 - 1 ---------ACAUGCAAUUUCACACAUGUGAGGAAAAUCACAAAACAGGCGUCGCAUUUUCAUAAUGGCAACAUAAA---AAAUGCACACA-------CAUACGUG---UA-UACAUAC ---------............((((.(((((......)))))......(.((.(((((((....(((....)))..)---)))))))).).-------.....)))---).-....... ( -19.50) >DroSim_CAF1 65352 96 - 1 ---------ACAUGCAAUUUCACACAUGUGAGGAAAAUCACAAAACAGGCGUCGCAUUUUCAUAAUGGCAACAUAAA---AAAUGCACACA-------CAUACGUG---UA-UACAUAC ---------............((((.(((((......)))))......(.((.(((((((....(((....)))..)---)))))))).).-------.....)))---).-....... ( -19.50) >DroEre_CAF1 66503 86 - 1 ---------ACAUGCAAUUUCACACAUGUGAGGAAAAUCACAAUACAGGCGUCGCAUUUUCAUAAUGGCAACAUAAA---AAAUGCAAACA-------CACAC--------------GC ---------....((...........(((((......)))))...........(((((((....(((....)))..)---)))))).....-------.....--------------)) ( -15.30) >DroYak_CAF1 65378 98 - 1 ---------ACAUGCAAUUUCACACAUGUGAGGAAAAUCACAAAACAGGCGUCGCAUUUUCAUAAUGGCAACAUAAAA--AAAUGCAAACA-------CAUACAUA---UAAUGCAUAC ---------..(((((..........(((((......)))))...........(((((((....(((....)))...)--)))))).....-------........---...))))).. ( -20.00) >DroAna_CAF1 64497 118 - 1 UACGUAUGUACAUGCAAUUUCACACAUGUGAGAAAAAACACAAAACAGACGUCGCACUCGCAUAAUGGCGGCAAAAAAAAAGAUGUAUAUAUUCUCCCCCUACAUGCUUUA-UAAUUUC ...((((((((((....((((((....)))))).................(((((.(.........))))))..........))))))))))...................-....... ( -21.10) >consensus _________ACAUGCAAUUUCACACAUGUGAGGAAAAUCACAAAACAGGCGUCGCAUUUUCAUAAUGGCAACAUAAA___AAAUGCAAACA_______CAUACGUG___UA_UACAUAC .................((((((....))))))....................((((((.....(((....)))......))))))................................. (-12.10 = -12.18 + 0.08)
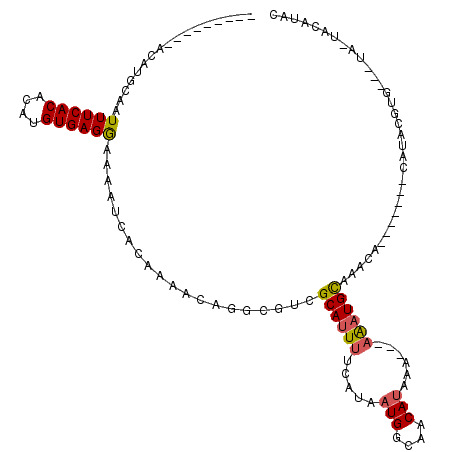
| Location | 8,758,388 – 8,758,480 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 84.52 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -21.69 |
| Energy contribution | -21.55 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8758388 92 + 22407834 --UUUAUGUUGCCAUUAUGAAAAUGCGACGCCUGUUUUGUGAUUUUCCUCACAUGUGUGAAAUUGCAUGU-------------------------AGGUGCGAUAGCGUUGAGGGGAAA --.....(((((.(((.....)))))))).............((..(((((.((((.....(((((((..-------------------------..))))))).)))))))))..)). ( -23.60) >DroSec_CAF1 62703 91 + 1 --UUUAUGUUGCCAUUAUGAAAAUGCGACGCCUGUUUUGUGAUUUUCCUCACAUGUGUGAAAUUGCAUGU-------------------------AGGUGCGAGAGCGUUGAGGGGA-A --.....(((((.(((.....)))))))).............((..(((((((((((......)))))))-------------------------....((....))...))))..)-) ( -23.90) >DroSim_CAF1 65380 92 + 1 --UUUAUGUUGCCAUUAUGAAAAUGCGACGCCUGUUUUGUGAUUUUCCUCACAUGUGUGAAAUUGCAUGU-------------------------AGGUGCGAGAGCGUUGAGGGGAAA --.....(((((.(((.....)))))))).............((..(((((((((((......)))))))-------------------------....((....))...))))..)). ( -24.70) >DroEre_CAF1 66521 92 + 1 --UUUAUGUUGCCAUUAUGAAAAUGCGACGCCUGUAUUGUGAUUUUCCUCACAUGUGUGAAAUUGCAUGU-------------------------AGGUGCGAGAGCAUUGAGGGGAAA --.....(((((.(((.....)))))))).............((..(((((((((((......)))))))-------------------------.(((((....)))))))))..)). ( -27.50) >DroYak_CAF1 65407 93 + 1 -UUUUAUGUUGCCAUUAUGAAAAUGCGACGCCUGUUUUGUGAUUUUCCUCACAUGUGUGAAAUUGCAUGU-------------------------AGGUGCGAGAGCAUUGAGGGGAAA -......(((((.(((.....)))))))).............((..(((((((((((......)))))))-------------------------.(((((....)))))))))..)). ( -27.50) >DroAna_CAF1 64536 117 + 1 UUUUUUUGCCGCCAUUAUGCGAGUGCGACGUCUGUUUUGUGUUUUUUCUCACAUGUGUGAAAUUGCAUGUACAUACGUACUACGUACUCGCACAUAGGUGCGAGCACAUUGAGGUGG-- ........(((((....((((((((((((((.(((..(((((..((((.((....)).))))..))))).))).))))....))))))))))((...(((....)))..)).)))))-- ( -42.40) >consensus __UUUAUGUUGCCAUUAUGAAAAUGCGACGCCUGUUUUGUGAUUUUCCUCACAUGUGUGAAAUUGCAUGU_________________________AGGUGCGAGAGCAUUGAGGGGAAA .......(((((.(((.....)))))))).............((..(((((((((((......)))))))..........................(((((....)))))))))..)). (-21.69 = -21.55 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:27 2006