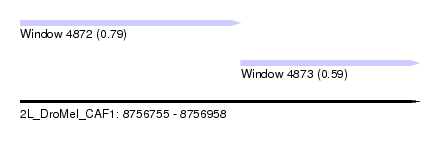
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,756,755 – 8,756,958 |
| Length | 203 |
| Max. P | 0.787780 |

| Location | 8,756,755 – 8,756,867 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 86.64 |
| Mean single sequence MFE | -18.78 |
| Consensus MFE | -16.96 |
| Energy contribution | -16.80 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
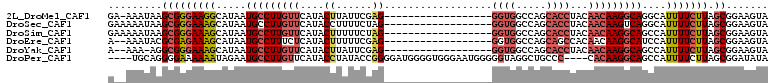
>2L_DroMel_CAF1 8756755 112 + 22407834 CGCAAAUAAUAAUAAGCAAGUUGGA-----UUCCAGAGCACGGGACUCGUUAACUCGCCUCC-GAAAGUUAAGAAAUGCAUAAAAUUUGUUAACAAGCCAAAAAAAAAGGAAAAAAAU .((............((.(((((((-----.(((........))).))..))))).))....-....(((((.((((.......)))).)))))..)).................... ( -16.20) >DroPse_CAF1 81019 108 + 1 CGCAAAUAAUAAUAAGCAAGUUGGAUGCCAUUCCAGAGACCGGGACUCGUUAACUUGCCUCCCAAAAGUUAAGAAAUGCAUAAAAUUUGUUAACAAGCUAAAAAAUAA---------- .((............((((((((((......))).(((.......)))...))))))).........(((((.((((.......)))).)))))..))..........---------- ( -21.30) >DroSim_CAF1 63766 107 + 1 CGCAAAUAAUAAUAAGCAAGUUGGA-----UUCCAGAGCACGGGACUCGUUAACUCGCCUCC-GAAAGUUAAGAAAUGCAUAAAAUUUGUUAACAAGCCAAAAAAU-----AAAAAAU .((............((.(((((((-----.(((........))).))..))))).))....-....(((((.((((.......)))).)))))..))........-----....... ( -16.20) >DroAna_CAF1 61431 102 + 1 CGCAAAUAAUAAUAAGCAAGUUGAA-----UUCCAGAGCUCGGGACUCGUUAACUCGCGUCC-AAAAGUUAAGAAAUGCAUAAAAUUUGUUAACAAGCAUAAAAAAAA---------- .((............((.((((((.-----.(((.(....).)))....)))))).))....-....(((((.((((.......)))).)))))..))..........---------- ( -18.90) >DroPer_CAF1 77379 108 + 1 CGCAAAUAAUAAUAAGCAAGUUGGAUGCCAUUCCAGAGACCGGGACUCGUUAACUUGCCUCCCAAAAGUUAAGAAAUGCAUAAAAUUUGUUAACAAGCUAAAAAAUAA---------- .((............((((((((((......))).(((.......)))...))))))).........(((((.((((.......)))).)))))..))..........---------- ( -21.30) >consensus CGCAAAUAAUAAUAAGCAAGUUGGA_____UUCCAGAGCACGGGACUCGUUAACUCGCCUCC_AAAAGUUAAGAAAUGCAUAAAAUUUGUUAACAAGCCAAAAAAUAA__________ .((............((.((((((......((((.......))))....)))))).)).........(((((.((((.......)))).)))))..)).................... (-16.96 = -16.80 + -0.16)


| Location | 8,756,867 – 8,756,958 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.03 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -14.27 |
| Energy contribution | -15.38 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8756867 91 + 22407834 GA-AAAUAAGCGGGAAGGCAUAAUGCCUUGUUCAUACUUAUUCGAG------------------GGUGGCCAGCACCUACAACAAGGCAGGCAUUUUCUUAGCGGAAGUA ..-........(..((.((....(((((((((....(((....)))------------------((((.....))))...))))))))).)).))..)...((....)). ( -25.80) >DroSec_CAF1 61207 92 + 1 GAAAAAUAAGCGGGAAAGCAUAAUGCCUUGUUCAUACCUUUUCUAG------------------GGUGGCCAGCACCUACAACAAGUCAGGCAUUUUCUUAGCGGAAGUA (((((....((......))....(((((((((..((((((....))------------------))))...)))).............))))))))))...((....)). ( -19.11) >DroSim_CAF1 63873 92 + 1 GAAAAAUAAGCGGGAAAGCAUAAUGCCUUGUUCAUACUUUUUCUAG------------------GGUGGCCAGCACCUACAACAAGGCAGCCAUUUUCUUAGCGGAAGUA (((((....((......))....(((((((((....((......))------------------((((.....))))...)))))))))....)))))...((....)). ( -23.80) >DroEre_CAF1 65029 90 + 1 A--AAAUACGCGAGAAAGCAUAAUGCCUUUCUCAUACUUUUUCGAG------------------GGUGGCCAGCAGCCACAACAAGGCAUCCAUUUUCUUAGCGGAAGUA .--.....((((((((((....(((((((((((..........)))------------------)(((((.....)))))...)))))))...))))))).)))...... ( -27.10) >DroYak_CAF1 63788 89 + 1 A--AAA-AGGCGGGAAAGCAUAAUGCCUUGUUCAUACUUAUUCGAG------------------GGUGGCCAGCACCUACAACAAGGCAGCCAUUUUCUUAGCGGAAGUA .--...-....(((((((.....(((((((((....(((....)))------------------((((.....))))...)))))))))....))))))).((....)). ( -24.10) >DroPer_CAF1 77487 102 + 1 ----UGCAGGGGAAAAAAUAGAAUGCCUUGUUCAUACCUAUACCGGGGAUGGGGUGGGAAUGGGGGUAGGCUGCCC----CACAAGGCAGCCAUUUUCUUAGCGGAUAUA ----.((..((((((..........(((..(((.(((((...(....)...))))).)))..)))...(((((((.----.....))))))).))))))..))....... ( -38.60) >consensus G__AAAUAAGCGGGAAAGCAUAAUGCCUUGUUCAUACUUAUUCGAG__________________GGUGGCCAGCACCUACAACAAGGCAGCCAUUUUCUUAGCGGAAGUA .........(((((((((.....(((((((((....((......))..................((((.....))))...)))))))))....))))))).))....... (-14.27 = -15.38 + 1.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:25 2006