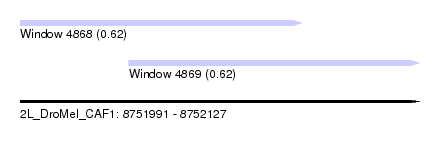
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,751,991 – 8,752,127 |
| Length | 136 |
| Max. P | 0.624882 |

| Location | 8,751,991 – 8,752,087 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 84.11 |
| Mean single sequence MFE | -29.87 |
| Consensus MFE | -19.70 |
| Energy contribution | -22.65 |
| Covariance contribution | 2.95 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8751991 96 + 22407834 UUUCUCGGGCUUCGGCUCUCUGAUUUCAUUU-UUUGUUUUCUUCUC-CGAAAGAGGA-------UGAGAGGGCCUCAAUUUGAUGGCCCAAAAACGAUGAAGAAU ......((((....)))).....(((((((.-(((..((((.((..-(....)..))-------.))))(((((((.....)).)))))..))).)))))))... ( -32.10) >DroSec_CAF1 56230 96 + 1 UUUCUCGGGCUUCGGCUCUCUGAUUUCAUUU-UUUGUUUUCUUCUC-CGAAAGAGGA-------UGAGUGGGCCUCAAAUUGAUGGCCCAAAAACGAUGAAGAAU ......((((....)))).....(((((((.-(((...(((.((..-(....)..))-------.)))((((((((.....)).)))))).))).)))))))... ( -31.30) >DroSim_CAF1 59288 96 + 1 UUUCUCGGGCUUCGGCUCUCUGAUUUCAUUU-UUUGUUUUCUUCUC-CGAAAGAGGA-------UGAGUGGGCCUCAAAUUGAUGGCCCAAAAACGAUGAAGAAU ......((((....)))).....(((((((.-(((...(((.((..-(....)..))-------.)))((((((((.....)).)))))).))).)))))))... ( -31.30) >DroEre_CAF1 60119 95 + 1 CUUCACGGGCUUCGGCUCCCUGAUUUCAUUU-UUUGUUUUCUUCUC-CGAAGCAGGA-------UGAGUGGGCCUCAAAUUGAUGGCCCAA-AACGAUGAAGAAU ..(((.((((....)).)).)))(((((((.-(((.....(((.((-(......)))-------.)))((((((((.....)).)))))))-)).)))))))... ( -31.80) >DroYak_CAF1 58643 103 + 1 CUUCUCGGGCUUCGGCUCUCUGAUUUCAUUU-UUUGUUUUCUUCUC-CAAUGGAGGAUUGAGGCUGAGUGGGCCUCAAAUUGAUGGCCCAAAAACGAUGAGGAAU (((((((...((((((.(((.((...((...-..))...))(((((-(...))))))..)))))))))((((((((.....)).))))))....))).))))... ( -35.00) >DroAna_CAF1 55690 92 + 1 UUUUUUUGGCUUCAGCUCCCUGUUUCCAUUUUUUUGUUUUCCUCUCUCCAAGGAGGC-------UCAAUCGACCUCUAUUCGAUGGACCGAAAAUGAGG------ ......(((...(((....)))...)))............((((.......(((((.-------((....))))))).((((......))))...))))------ ( -17.70) >consensus UUUCUCGGGCUUCGGCUCUCUGAUUUCAUUU_UUUGUUUUCUUCUC_CGAAAGAGGA_______UGAGUGGGCCUCAAAUUGAUGGCCCAAAAACGAUGAAGAAU ((((..((((....))))...............((((((((((((......)))))............((((((((.....)).))))))))))))).))))... (-19.70 = -22.65 + 2.95)
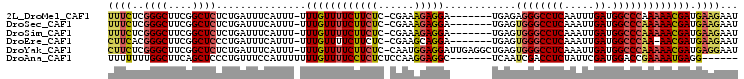
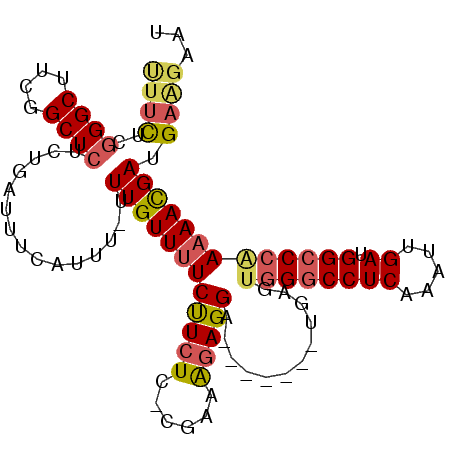
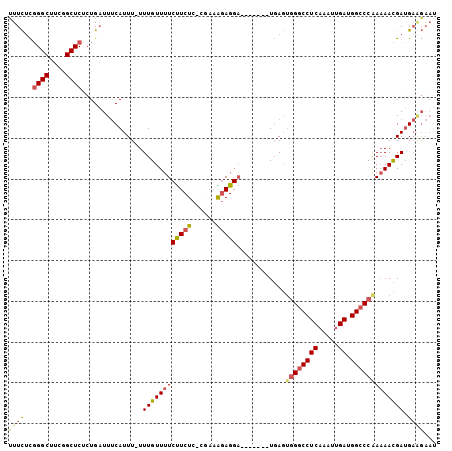
| Location | 8,752,028 – 8,752,127 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.70 |
| Mean single sequence MFE | -31.83 |
| Consensus MFE | -22.71 |
| Energy contribution | -24.38 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8752028 99 + 22407834 UUCUUCUCCGAAAGAGGA-------UGAGAGGGCCUCAAUUUGAUGGCCCAAAA------------ACGAUGAAGAAU-AAGCCCUCAAGAGGUGUGGCAGAGUCGGCACUUUUGCGGC (((((((((......)))-------.....(((((((.....)).)))))....------------.....)))))).-..(((....((((((((.(......).))))))))..))) ( -32.00) >DroSec_CAF1 56267 99 + 1 UUCUUCUCCGAAAGAGGA-------UGAGUGGGCCUCAAAUUGAUGGCCCAAAA------------ACGAUGAAGAAU-AAGCCCUCAAGAGGUGUGGCAGAGUCGGCACUUUUGCGGC (((((((((......)))-------....((((((((.....)).))))))...------------.....)))))).-..(((....((((((((.(......).))))))))..))) ( -32.70) >DroSim_CAF1 59325 99 + 1 UUCUUCUCCGAAAGAGGA-------UGAGUGGGCCUCAAAUUGAUGGCCCAAAA------------ACGAUGAAGAAU-AAGCCCUCAAGAGGUGUGGCAGAGUCGGCACUUUUGCGGC (((((((((......)))-------....((((((((.....)).))))))...------------.....)))))).-..(((....((((((((.(......).))))))))..))) ( -32.70) >DroEre_CAF1 60156 98 + 1 UUCUUCUCCGAAGCAGGA-------UGAGUGGGCCUCAAAUUGAUGGCCCAA-A------------ACGAUGAAGAAU-AAGCCCUCAAGAGGUGUGGCAGAGUCGGCACUUUUGCGGC (((((((((......)))-------....((((((((.....)).)))))).-.------------.....)))))).-..(((....((((((((.(......).))))))))..))) ( -32.70) >DroYak_CAF1 58680 106 + 1 UUCUUCUCCAAUGGAGGAUUGAGGCUGAGUGGGCCUCAAAUUGAUGGCCCAAAA------------ACGAUGAGGAAU-AAGUCCUCAGGAGGUGUGGCAGAGUCGGCACUUUUGCGGC ...((((((...)))))).....((((..((((((((.....)).))))))...------------....((((((..-...))))))((((((((.(......).)))))))).)))) ( -36.60) >DroPer_CAF1 71446 99 + 1 U-C------------GGC-------UCAGUCGGCCUCAAUUUGAUUUCCAAAAAAAAUGGCAAAAGAAGAUGAAGAAUCGAGCGGUCUUGAGGUGUGGCACAGUCGGCACUUUUGUGGU (-(------------(((-------(..(((((((((((((((.....))))......(((....(..(((.....)))...).)))))))))).))))..))))))(((....))).. ( -24.30) >consensus UUCUUCUCCGAAAGAGGA_______UGAGUGGGCCUCAAAUUGAUGGCCCAAAA____________ACGAUGAAGAAU_AAGCCCUCAAGAGGUGUGGCAGAGUCGGCACUUUUGCGGC (((((((((......)))...........((((((((.....)).))))))....................))))))....(((....((((((((.(......).))))))))..))) (-22.71 = -24.38 + 1.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:21 2006