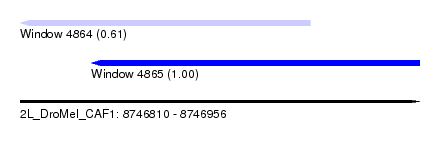
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,746,810 – 8,746,956 |
| Length | 146 |
| Max. P | 0.999698 |

| Location | 8,746,810 – 8,746,916 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.01 |
| Mean single sequence MFE | -18.88 |
| Consensus MFE | -18.57 |
| Energy contribution | -18.02 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.606946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
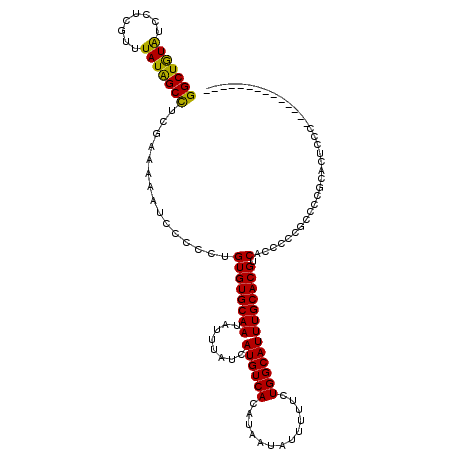
>2L_DroMel_CAF1 8746810 106 - 22407834 GGCUGUAUCCUCGUUUAUAGCCUCGAAAAAUCCCCCUGUGUGCAAUAUUUAUCAUGUCACAUAAUAUUUUUCUGGCAUUUGCACGUCACCCCCGCCCCGUACUCCU-------------- (((((((........)))))))..............(((((((((........((((((.............))))))))))))).))..................-------------- ( -19.22) >DroSec_CAF1 50990 106 - 1 GGCUGUAUCCUCGUUUAUAGCCUCGAAAAAUCCCCCUGUGUGCAAUAUUUAUCAUGUCACAUAAUAUUUUUCUGGCAUUUGCACGUCACCCCCGCACCGCACUCUC-------------- (((((((........)))))))..(((((((.....((((((((..........)).))))))..)))))))..((...(((...........)))..))......-------------- ( -19.50) >DroSim_CAF1 54118 106 - 1 GGCUGUAUCCUCGUUUAUAGCCUCGAAAAAUCCCCCUGUGUGCAAUAUUUAUCAUGUCACAUAAUAUUUUUCUGGCAUUUGCACGUCACCCCCGCCCCGCACUCUC-------------- (((((((........)))))))..............(((((((((........((((((.............))))))))))))).))..................-------------- ( -19.22) >DroEre_CAF1 54926 108 - 1 GGCUGUAUCCCCGUUUAUGGCCUCGAAAAAUCCCCCUGUGUGCAAUAUUUAUCAUGUCACAUAAUAUUUUUCUGGCAUUUGCACGUCACCC-CGCCCAGCACUCCU-----------CCC .((((.......((..(((.((..(((((((.....((((((((..........)).))))))..))))))).)))))..)).((......-))..))))......-----------... ( -18.60) >DroYak_CAF1 53384 99 - 1 GGCUGUAUCCUCGUUUAUAGCCUCGAAAAAUCCCCCUGUGUGCAAUAUUUAUCAUGUCACAUAAUAUUUUUCUGGCAUUUGCACGGCACUC-CC---------CCG-----------UCC (((((((........)))))))..(((((((.....((((((((..........)).))))))..)))))))..((....))((((.....-..---------)))-----------).. ( -20.90) >DroAna_CAF1 50523 114 - 1 GGCUAUGUCUACGUUUAUAGCUUCGAA-AAUCCCCCAGUGUGCAAUAUUUAUCAUGUCACAUAAUAUUUUUCUGGCAUUUGCACGUCACCUCCAC-----ACUCUCUGCCCUCCACUCGC (((((((........))))))).....-.........((((((((........((((((.............))))))))))))).)........-----.................... ( -15.82) >consensus GGCUGUAUCCUCGUUUAUAGCCUCGAAAAAUCCCCCUGUGUGCAAUAUUUAUCAUGUCACAUAAUAUUUUUCUGGCAUUUGCACGUCACCCCCGCCCCGCACUCCC______________ (((((((........)))))))...............((((((((........((((((.............))))))))))))).)................................. (-18.57 = -18.02 + -0.55)


| Location | 8,746,836 – 8,746,956 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.72 |
| Mean single sequence MFE | -25.83 |
| Consensus MFE | -25.04 |
| Energy contribution | -24.98 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.91 |
| SVM RNA-class probability | 0.999698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
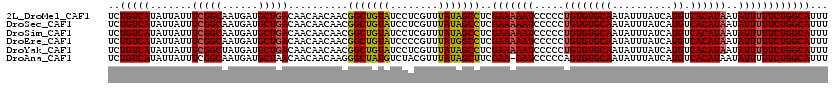
>2L_DroMel_CAF1 8746836 120 - 22407834 UCUGUCAUAUUAUUUCGGCAAUGAUGCUGACAACAACAACGGCUGUAUCCUCGUUUAUAGCCUCGAAAAAUCCCCCUGUGUGCAAUAUUUAUCAUGUCACAUAAUAUUUUUCUGGCAUUU ..(((((.......((((((....))))))..........(((((((........)))))))..(((((((.....((((((((..........)).))))))..))))))))))))... ( -27.20) >DroSec_CAF1 51016 120 - 1 UCUGUCAUAUUAUUUCGGCAAUGAUGCUGACAACAACAACGGCUGUAUCCUCGUUUAUAGCCUCGAAAAAUCCCCCUGUGUGCAAUAUUUAUCAUGUCACAUAAUAUUUUUCUGGCAUUU ..(((((.......((((((....))))))..........(((((((........)))))))..(((((((.....((((((((..........)).))))))..))))))))))))... ( -27.20) >DroSim_CAF1 54144 120 - 1 UCUGUCAUAUUAUUUCGGCAAUGAUGCUGACAACAACAACGGCUGUAUCCUCGUUUAUAGCCUCGAAAAAUCCCCCUGUGUGCAAUAUUUAUCAUGUCACAUAAUAUUUUUCUGGCAUUU ..(((((.......((((((....))))))..........(((((((........)))))))..(((((((.....((((((((..........)).))))))..))))))))))))... ( -27.20) >DroEre_CAF1 54954 120 - 1 UCUGUCAUAUUAUUUCGGCAAUGAUGCUGACAACAACAACGGCUGUAUCCCCGUUUAUGGCCUCGAAAAAUCCCCCUGUGUGCAAUAUUUAUCAUGUCACAUAAUAUUUUUCUGGCAUUU ..(((((.......((((((....))))))..........(((((((........)))))))..(((((((.....((((((((..........)).))))))..))))))))))))... ( -25.90) >DroYak_CAF1 53403 120 - 1 UCUGUCAUAUUAUUUCGGCUAUGAUGCUGACAACAACAACGGCUGUAUCCUCGUUUAUAGCCUCGAAAAAUCCCCCUGUGUGCAAUAUUUAUCAUGUCACAUAAUAUUUUUCUGGCAUUU ..(((((.......(((((......)))))..........(((((((........)))))))..(((((((.....((((((((..........)).))))))..))))))))))))... ( -27.00) >DroAna_CAF1 50558 119 - 1 UCUGUCAUAUUAUUUCGGCAAUGAUGCUAACAACAACAAGGGCUAUGUCUACGUUUAUAGCUUCGAA-AAUCCCCCAGUGUGCAAUAUUUAUCAUGUCACAUAAUAUUUUUCUGGCAUUU ..((((..........))))..(((((((..........((((((((........)))))))).(((-((.......(((((((..........)).))))).....)))))))))))). ( -20.50) >consensus UCUGUCAUAUUAUUUCGGCAAUGAUGCUGACAACAACAACGGCUGUAUCCUCGUUUAUAGCCUCGAAAAAUCCCCCUGUGUGCAAUAUUUAUCAUGUCACAUAAUAUUUUUCUGGCAUUU ..(((((.......(((((......)))))..........(((((((........)))))))..(((((((.....((((((((..........)).))))))..))))))))))))... (-25.04 = -24.98 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:17 2006