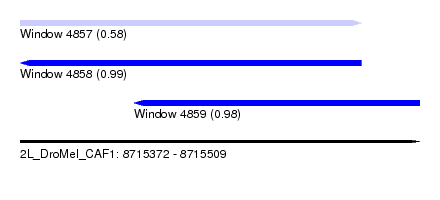
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,715,372 – 8,715,509 |
| Length | 137 |
| Max. P | 0.987092 |

| Location | 8,715,372 – 8,715,489 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 85.22 |
| Mean single sequence MFE | -30.26 |
| Consensus MFE | -19.76 |
| Energy contribution | -20.68 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8715372 117 + 22407834 UAAAUGCAGAUUAUCUUUUUGUCAUGAUUGUAUGUUC-CUUGAGUAUUCCAAGUCUAUGCUCCCAAUUUCUGGCAACACAGACUAGCUGGCUGUGAAACAACUCAAGACCAAGUGCAG ...(((((((.......)))).)))...(((((....-(((((((.(((..((((...(((.......((((......))))..))).))))..)))...))))))).....))))). ( -24.10) >DroSec_CAF1 27940 117 + 1 UAACUGCAGACUAUCUAUUUGUCAAGAUUGUCUGUUC-CUUGAGUAUUCCAAGUCUGUGCUCCCAAUUUCUGGCAACACAGACUAGCUGGCUAUGAAACAACUCAAGACCAAGUGCAG ...((((((((.((((........)))).))))....-(((((((......((((((((...(((.....)))...))))))))((....))........))))))).......)))) ( -34.20) >DroSim_CAF1 28648 117 + 1 UAACUGCAGACUAUCUAUUUGUCAAGAUUGUCUGUUC-CUUGAGUAUUCCAAGUCUGUGCUCCCAAUUUCUGGCAACACAGACUAGCUGGCUAUGAAACAACUCAAGACCAAGUGCAG ...((((((((.((((........)))).))))....-(((((((......((((((((...(((.....)))...))))))))((....))........))))))).......)))) ( -34.20) >DroEre_CAF1 30369 113 + 1 UAAAUACAGACUAUCAUUCC-----UGUUGCAUGUUUUCUUAGCUGCUCCAAGUCUGUGCUCCCAAUUUCUGGCAACACAGACUAGUUGGCUGUGGAACAACUCAAGACCAAGUGCAG .....((((..........)-----)))(((((....((((((.((.((((((((((((...(((.....)))...))))))))((....)).)))).)).)).))))....))))). ( -29.80) >DroYak_CAF1 28227 118 + 1 UAAGUGGAGACUAUCAUUCCUUCAAUGUUGUAUGAUUUCUGAUAUACUCCAAGUCUGUGCUCCCAAUUUCUGGCUACGCAGACAAGUUGGCUGUGAAACAACUCAAGACCAAGUGCAG ....(((((..(((((.....(((........)))....)))))..))))).(((((((...(((.....)))...))))))).(.((((((.(((......))))).)))).).... ( -29.00) >consensus UAAAUGCAGACUAUCUUUUUGUCAAGAUUGUAUGUUC_CUUGAGUAUUCCAAGUCUGUGCUCCCAAUUUCUGGCAACACAGACUAGCUGGCUGUGAAACAACUCAAGACCAAGUGCAG ............................(((((.....(((((((((.(((((((((((...(((.....)))...))))))))...)))..))......))))))).....))))). (-19.76 = -20.68 + 0.92)



| Location | 8,715,372 – 8,715,489 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 85.22 |
| Mean single sequence MFE | -39.42 |
| Consensus MFE | -25.88 |
| Energy contribution | -26.32 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.68 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8715372 117 - 22407834 CUGCACUUGGUCUUGAGUUGUUUCACAGCCAGCUAGUCUGUGUUGCCAGAAAUUGGGAGCAUAGACUUGGAAUACUCAAG-GAACAUACAAUCAUGACAAAAAGAUAAUCUGCAUUUA .((((.....((((((((.((((((.((....))((((((((((.((((...)))).)))))))))))))))))))))))-)..(((......)))..............)))).... ( -33.20) >DroSec_CAF1 27940 117 - 1 CUGCACUUGGUCUUGAGUUGUUUCAUAGCCAGCUAGUCUGUGUUGCCAGAAAUUGGGAGCACAGACUUGGAAUACUCAAG-GAACAGACAAUCUUGACAAAUAGAUAGUCUGCAGUUA ((((......((((((((.(((((((((....)))(((((((((.((((...)))).))))))))).)))))))))))))-)...((((.((((........)))).))))))))... ( -45.30) >DroSim_CAF1 28648 117 - 1 CUGCACUUGGUCUUGAGUUGUUUCAUAGCCAGCUAGUCUGUGUUGCCAGAAAUUGGGAGCACAGACUUGGAAUACUCAAG-GAACAGACAAUCUUGACAAAUAGAUAGUCUGCAGUUA ((((......((((((((.(((((((((....)))(((((((((.((((...)))).))))))))).)))))))))))))-)...((((.((((........)))).))))))))... ( -45.30) >DroEre_CAF1 30369 113 - 1 CUGCACUUGGUCUUGAGUUGUUCCACAGCCAACUAGUCUGUGUUGCCAGAAAUUGGGAGCACAGACUUGGAGCAGCUAAGAAAACAUGCAACA-----GGAAUGAUAGUCUGUAUUUA .((((..(..((((.((((((((((.........((((((((((.((((...)))).))))))))))))))))))))))))..)..))))(((-----((........)))))..... ( -42.80) >DroYak_CAF1 28227 118 - 1 CUGCACUUGGUCUUGAGUUGUUUCACAGCCAACUUGUCUGCGUAGCCAGAAAUUGGGAGCACAGACUUGGAGUAUAUCAGAAAUCAUACAACAUUGAAGGAAUGAUAGUCUCCACUUA ......(((((..((((....))))..)))))...(((((.((..((((...))))..)).))))).(((((..(((((....(((........))).....)))))..))))).... ( -30.50) >consensus CUGCACUUGGUCUUGAGUUGUUUCACAGCCAGCUAGUCUGUGUUGCCAGAAAUUGGGAGCACAGACUUGGAAUACUCAAG_GAACAUACAAUCUUGACAAAAAGAUAGUCUGCAGUUA .((((.(((((..((((....))))..)))))(.((((((((((.((((...)))).)))))))))).).........................................)))).... (-25.88 = -26.32 + 0.44)


| Location | 8,715,411 – 8,715,509 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 92.45 |
| Mean single sequence MFE | -31.64 |
| Consensus MFE | -28.34 |
| Energy contribution | -28.82 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8715411 98 - 22407834 UAGACAAGUUUGACCGCCGUCUGCACUUGGUCUUGAGUUGUUUCACAGCCAGCUAGUCUGUGUUGCCAGAAAUUGGGAGCAUAGACUUGGAAUACUCA .((((((((..(((....)))...)))).))))(((((.((((((.((....))((((((((((.((((...)))).))))))))))))))))))))) ( -32.50) >DroSec_CAF1 27979 98 - 1 UAGACAAGUUUGACCGCCGCCUGCACUUGGUCUUGAGUUGUUUCAUAGCCAGCUAGUCUGUGUUGCCAGAAAUUGGGAGCACAGACUUGGAAUACUCA ...........(((((..(....)...))))).(((((.(((((((((....)))(((((((((.((((...)))).))))))))).))))))))))) ( -32.10) >DroSim_CAF1 28687 98 - 1 UAGACAAGUUUGACCGCCGCCUGCACUUGGUCUUGAGUUGUUUCAUAGCCAGCUAGUCUGUGUUGCCAGAAAUUGGGAGCACAGACUUGGAAUACUCA ...........(((((..(....)...))))).(((((.(((((((((....)))(((((((((.((((...)))).))))))))).))))))))))) ( -32.10) >DroEre_CAF1 30404 98 - 1 UAGACGAGUUUGACCGCCGCCUGCACUUGGUCUUGAGUUGUUCCACAGCCAACUAGUCUGUGUUGCCAGAAAUUGGGAGCACAGACUUGGAGCAGCUA ....((((.(..(..((.....))..)..).))))((((((((((.........((((((((((.((((...)))).)))))))))))))))))))). ( -36.80) >DroYak_CAF1 28267 98 - 1 UAGACAAGUUUGACCGCCGCCUGCACUUGGUCUUGAGUUGUUUCACAGCCAACUUGUCUGCGUAGCCAGAAAUUGGGAGCACAGACUUGGAGUAUAUC ....((((((((...(((((..(((.(((((..((((....))))..)))))..)))..)))...((((...))))..)).))))))))......... ( -24.70) >consensus UAGACAAGUUUGACCGCCGCCUGCACUUGGUCUUGAGUUGUUUCACAGCCAGCUAGUCUGUGUUGCCAGAAAUUGGGAGCACAGACUUGGAAUACUCA .((((((.((.(((((..(....)...)))))..)).)))))).....(((...((((((((((.((((...)))).)))))))))))))........ (-28.34 = -28.82 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:11 2006