
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,703,753 – 8,703,850 |
| Length | 97 |
| Max. P | 0.990618 |

| Location | 8,703,753 – 8,703,850 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 79.41 |
| Mean single sequence MFE | -22.53 |
| Consensus MFE | -15.59 |
| Energy contribution | -16.23 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
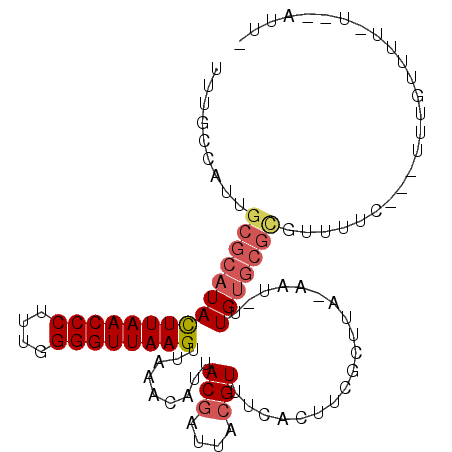
>2L_DroMel_CAF1 8703753 97 + 22407834 UUUGCCAUUGCGCAUACUUAACCCUUUGGGGUUAAGUUAAACAUUACGAUUACGUUUCACAUCGCUUA-AAU-UUGUGCGCGUUUCC---UUUGUUUU-U--ACU- .........(((((((((((((((....))))))))).........((((..........))))....-...-..))))))......---........-.--...- ( -22.80) >DroSec_CAF1 16295 99 + 1 UUUGCCAUUGAGCAUACUUAACCCUUUGGGGUUAAGUUAAACAUUACGAUUACGUUUCACAUCGCUUA-AAU-UUGUGCGUGUUUUU---UUUGUUUUUU--AUUU ...((((((((((..(((((((((....)))))))))........(((....)))........)))))-)..-.)).))........---..........--.... ( -20.70) >DroEre_CAF1 18860 94 + 1 UUUGCCAUUGCGCAUACUUAACCCUUUGGGGUUAAGUUAAACAUUACGAUUACGUUUCACGUCGCUUA-AAUGUUGUGCGCGUUUUC---UUUGUUUU-------- .........(((((((((((((((....)))))))))..((((((......(((.....)))......-))))))))))))......---........-------- ( -26.90) >DroWil_CAF1 19287 97 + 1 UUUGCUUUUGCGCAUAUUUAACCCUUUGGGGUUAAGUUAAGCAUUACGUUUAAGUUUUACUUCGCUUU-GUU-UUGUAAGGUUUUUA---UUUGACUUC---GUU- ...((....))((..(((((((((....)))))))))...))...(((.((((((...((((..(...-...-..)..))))....)---)))))...)---)).- ( -21.70) >DroMoj_CAF1 24619 97 + 1 UUUGCUAUUGCGCAUAUUUAACCCUUUAGGGUUAAGUUAAACAUUACGCUUACGUUUUAUUUCGUUUUCAUU-UUGCGCGCCAU-U-----UUGCGAUUU--AUUU .((((.((.((((..(((((((((....)))))))))..........((..(((........))).......-..)))))).))-.-----..))))...--.... ( -21.80) >DroPer_CAF1 19012 102 + 1 UUUGCUGUUGCGCAUAUUUAACCCUU--GGGUUAAGUUAAACAUUACGAUUACGUUUUACUUCGCUUA-AUU-UUGUGCGCGUUUUCCCCAUGGUUUUCCAUACAC .........((((((((((((((...--.)))))))...........(((((.((........)).))-)))-.))))))).........((((....)))).... ( -21.30) >consensus UUUGCCAUUGCGCAUACUUAACCCUUUGGGGUUAAGUUAAACAUUACGAUUACGUUUCACUUCGCUUA_AAU_UUGUGCGCGUUUUC___UUUGUUUU_U__AUU_ .........(((((((((((((((....)))))))).........(((....)))...................)))))))......................... (-15.59 = -16.23 + 0.64)


| Location | 8,703,753 – 8,703,850 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 79.41 |
| Mean single sequence MFE | -23.42 |
| Consensus MFE | -13.66 |
| Energy contribution | -14.55 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.10 |
| Mean z-score | -4.09 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8703753 97 - 22407834 -AGU--A-AAAACAAA---GGAAACGCGCACAA-AUU-UAAGCGAUGUGAAACGUAAUCGUAAUGUUUAACUUAACCCCAAAGGGUUAAGUAUGCGCAAUGGCAAA -.((--.-........---(....)(((((...-...-...(((((..........)))))........(((((((((....))))))))).)))))....))... ( -27.00) >DroSec_CAF1 16295 99 - 1 AAAU--AAAAAACAAA---AAAAACACGCACAA-AUU-UAAGCGAUGUGAAACGUAAUCGUAAUGUUUAACUUAACCCCAAAGGGUUAAGUAUGCUCAAUGGCAAA ....--...(((((..---....((((((....-...-...))).)))...(((....)))..))))).(((((((((....))))))))).((((....)))).. ( -22.30) >DroEre_CAF1 18860 94 - 1 --------AAAACAAA---GAAAACGCGCACAACAUU-UAAGCGACGUGAAACGUAAUCGUAAUGUUUAACUUAACCCCAAAGGGUUAAGUAUGCGCAAUGGCAAA --------........---......(((((.((((((-......(((.....)))......))))))..(((((((((....))))))))).)))))......... ( -26.80) >DroWil_CAF1 19287 97 - 1 -AAC---GAAGUCAAA---UAAAAACCUUACAA-AAC-AAAGCGAAGUAAAACUUAAACGUAAUGCUUAACUUAACCCCAAAGGGUUAAAUAUGCGCAAAAGCAAA -...---.........---..............-...-...((((((((..((......))..)))))...(((((((....))))))).....)))......... ( -14.60) >DroMoj_CAF1 24619 97 - 1 AAAU--AAAUCGCAA-----A-AUGGCGCGCAA-AAUGAAAACGAAAUAAAACGUAAGCGUAAUGUUUAACUUAACCCUAAAGGGUUAAAUAUGCGCAAUAGCAAA ....--.....((..-----.-...))(((((.-..........(((((..(((....)))..)))))...(((((((....)))))))...)))))......... ( -22.30) >DroPer_CAF1 19012 102 - 1 GUGUAUGGAAAACCAUGGGGAAAACGCGCACAA-AAU-UAAGCGAAGUAAAACGUAAUCGUAAUGUUUAACUUAACCC--AAGGGUUAAAUAUGCGCAACAGCAAA (((((((...((((...(((....(((......-...-...)))((((.((((((.......)))))).))))..)))--...))))...)))))))......... ( -27.50) >consensus _AAU__A_AAAACAAA___GAAAACGCGCACAA_AAU_UAAGCGAAGUAAAACGUAAUCGUAAUGUUUAACUUAACCCCAAAGGGUUAAAUAUGCGCAAUAGCAAA .........................(((((................((.((((((.......)))))).))(((((((....)))))))...)))))......... (-13.66 = -14.55 + 0.89)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:06 2006