
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,698,040 – 8,698,133 |
| Length | 93 |
| Max. P | 0.999469 |

| Location | 8,698,040 – 8,698,133 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 88.80 |
| Mean single sequence MFE | -26.94 |
| Consensus MFE | -22.77 |
| Energy contribution | -22.89 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.63 |
| SVM RNA-class probability | 0.999469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
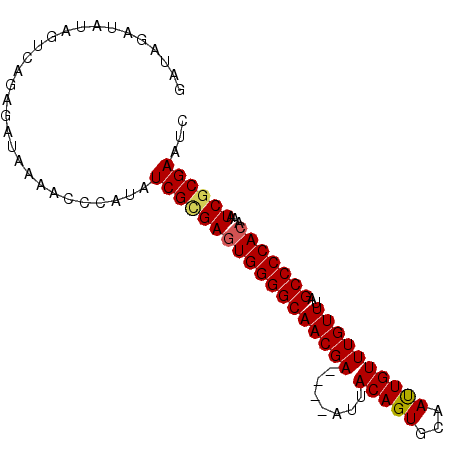
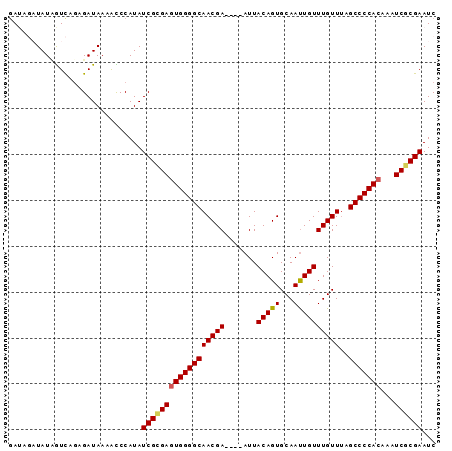
>2L_DroMel_CAF1 8698040 93 + 22407834 GAUAGAUAUCAUCAGAGAUAAAAUCCAUAUCGCGAGUGGGGCAACGAAUGAAUUACAGUGCAAUUGUUUGUUUAGCCCCACAAAUCGCGAAUC ......((((......)))).........(((((((((((((((((((..(............)..))))))..)))))))...))))))... ( -28.50) >DroSec_CAF1 10655 89 + 1 UAUAGAUAUUGUCAGAGAUAAAAUCCAUAUCGCGAGUGGGGCAACGA----AUUACAGUGCAAUUGUUUGUUAAGCCCCACAAAUCGCGAAUC (((.(((.(((((...))))).))).)))((((((((((((((((((----((............)))))))..)))))))...))))))... ( -29.60) >DroSim_CAF1 11423 89 + 1 GAUAGAUAUUGUCAGAGAUAAAACCCAUAUCGCGAGUGGGGCAACGA----AUUACAGUGCAAUUGUUUGUUUAGCCCCACAAAUCGCGAAUC (((((...)))))................((((((((((((((((((----((............)))))))..)))))))...))))))... ( -27.30) >DroEre_CAF1 11301 85 + 1 GA----UAUAGUUCAUGAUAAAGCCCAUAUCGGGAAUGGGGCAACGA----AUUACAGUGCAACUGUUUGUUUAGCCCCACAAAUCGCGAAUC ..----....((((.((((....(((.....)))..(((((((((((----...((((.....)))))))))..))))))...)))).)))). ( -24.80) >DroYak_CAF1 10859 85 + 1 GA----UAUAGUCAGAGAUAAAACCCAUAUCGUGAGUGGGGCAACGA----AUUACAGUGCAAUUGUUUGUUUAGCCCCACAAAUCGCGAAUC ..----.......................((((((((((((((((((----((............)))))))..)))))))...))))))... ( -24.50) >consensus GAUAGAUAUAGUCAGAGAUAAAACCCAUAUCGCGAGUGGGGCAACGA____AUUACAGUGCAAUUGUUUGUUUAGCCCCACAAAUCGCGAAUC .............................((((((((((((((((((.......(((((...))))))))))..)))))))...))))))... (-22.77 = -22.89 + 0.12)

| Location | 8,698,040 – 8,698,133 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 88.80 |
| Mean single sequence MFE | -25.24 |
| Consensus MFE | -21.27 |
| Energy contribution | -21.35 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.02 |
| SVM RNA-class probability | 0.998144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
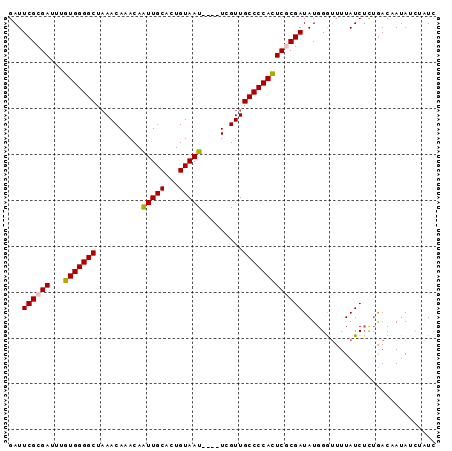
>2L_DroMel_CAF1 8698040 93 - 22407834 GAUUCGCGAUUUGUGGGGCUAAACAAACAAUUGCACUGUAAUUCAUUCGUUGCCCCACUCGCGAUAUGGAUUUUAUCUCUGAUGAUAUCUAUC ...((((((...(((((((.........((((((...))))))........))))))))))))).((((((.(((((...))))).)))))). ( -30.43) >DroSec_CAF1 10655 89 - 1 GAUUCGCGAUUUGUGGGGCUUAACAAACAAUUGCACUGUAAU----UCGUUGCCCCACUCGCGAUAUGGAUUUUAUCUCUGACAAUAUCUAUA ...((((((...(((((((......(((((((((...)))))----).))))))))))))))))(((((((.((.((...)).)).))))))) ( -27.60) >DroSim_CAF1 11423 89 - 1 GAUUCGCGAUUUGUGGGGCUAAACAAACAAUUGCACUGUAAU----UCGUUGCCCCACUCGCGAUAUGGGUUUUAUCUCUGACAAUAUCUAUC .....((((...(((((((......(((((((((...)))))----).))))))))))))))(((((..(((........))).))))).... ( -26.20) >DroEre_CAF1 11301 85 - 1 GAUUCGCGAUUUGUGGGGCUAAACAAACAGUUGCACUGUAAU----UCGUUGCCCCAUUCCCGAUAUGGGCUUUAUCAUGAACUAUA----UC ..((((.(((..(((((((.......((((.....))))...----.....))))))).(((.....)))....))).)))).....----.. ( -21.06) >DroYak_CAF1 10859 85 - 1 GAUUCGCGAUUUGUGGGGCUAAACAAACAAUUGCACUGUAAU----UCGUUGCCCCACUCACGAUAUGGGUUUUAUCUCUGACUAUA----UC ............(((((((......(((((((((...)))))----).))))))))))....(((((((.............)))))----)) ( -20.92) >consensus GAUUCGCGAUUUGUGGGGCUAAACAAACAAUUGCACUGUAAU____UCGUUGCCCCACUCGCGAUAUGGGUUUUAUCUCUGACAAUAUCUAUC ...((((((...(((((((..........(((((...))))).........)))))))))))))............................. (-21.27 = -21.35 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:02 2006