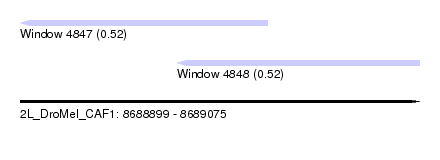
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,688,899 – 8,689,075 |
| Length | 176 |
| Max. P | 0.524895 |

| Location | 8,688,899 – 8,689,008 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.40 |
| Mean single sequence MFE | -30.95 |
| Consensus MFE | -19.36 |
| Energy contribution | -19.37 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8688899 109 - 22407834 AUCGAUGAACUGGUGACGGUCAUGAAGGAUGUGGGUAUCGAUGACACUGAAUUCGCUUGCAUCAAGGCCCUAGUCUUCUUCGAUCCCAGUGAGUUAUUUAACUUAUUUA ((((((((.(((....)))))).(((((....((((...((((....((....))....))))...))))...))))).)))))...((((((((....)))))))).. ( -30.10) >DroGri_CAF1 1945 99 - 1 AUUGAUGAGCUGGUGGUCGUCAUGAAGGAUGUCGGCAUUGAUGAGACCGAAUUUGCCUGCAUCAAGGCGCUGGUCUUCUUUGACCCCAGUGAGUUAA-----AU----- .(((((..(((((.(((((....(((((...(((((.(((.......)))....((((......))))))))))))))..))))))))))..)))))-----..----- ( -34.90) >DroEre_CAF1 1677 109 - 1 AUCGAUGAACUGGUGACGGUCAUGAAGGAUGUGGGCAUCGAUGAUACUGAAUUCGCCUGCAUCAAGGCCCUAGUCUUCUUCGAUCCCAGUGAGUUAUUCAACCUAUUGA .((((((.((((....))))..((((.(((.((((.(((((.((.((((.....((((......))))..))))..)).)))))))))....))).))))...)))))) ( -31.40) >DroWil_CAF1 1130 109 - 1 AUUGAUGAACUGGUUGUGGUGAUGAAAGAUGUGGGCAUUGACGAUACGGAAUUCGCCUGCAUUAAGGCGUUGGUGUUCUUUGAUCCAAGUGAGUAGAUCUCGAUUGUGA (((((.((.(((.(..(...(((.(((((.....((((..((((........))((((......))))))..))))))))).)))...)..).))).)))))))..... ( -30.20) >DroYak_CAF1 1275 109 - 1 AUCGAUGAACUGGUGACGGUUAUGAAGGAUGUGGGCAUCGAUGAUACGGAAUUCGCUUGCAUCAAGGCCCUAGUCUUCUUUGAUCCUAGUAGGUUAUUCGACAUAGUUA .((((..(((((....)))))..(((((....((((...((((...((.....))....))))...))))...)))))..(((((......))))).))))........ ( -28.50) >DroMoj_CAF1 1662 104 - 1 AUUGAUGAACUGGUGGUCGUUAUGAAGGACGUGGGCAUCGAUGAUACCGAAUUUGCCUGUAUUAAGGCAUUGGUCUUCUUUGAUCCCAGUAAGUGGUGUCGAUU----- (((((((.(((....(((.(.....).))).((((.(((((.((.(((((...(((((......))))))))))..)).)))))))))...)))..))))))).----- ( -30.60) >consensus AUCGAUGAACUGGUGACGGUCAUGAAGGAUGUGGGCAUCGAUGAUACCGAAUUCGCCUGCAUCAAGGCCCUAGUCUUCUUUGAUCCCAGUGAGUUAUUCAACAUA_U_A ...(((..(((((....((((..((((((((..(((..................)))..))))(((((....))))))))))))))))))..))).............. (-19.36 = -19.37 + 0.01)

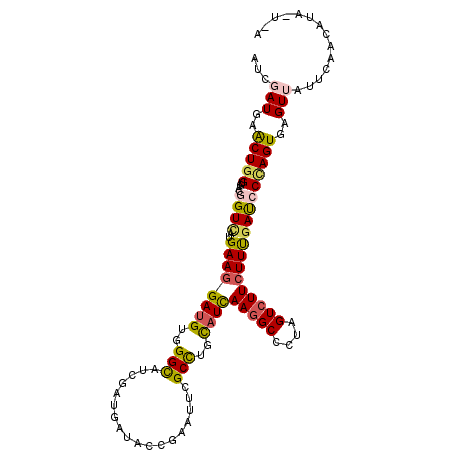

| Location | 8,688,968 – 8,689,075 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.55 |
| Mean single sequence MFE | -34.20 |
| Consensus MFE | -21.96 |
| Energy contribution | -22.63 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8688968 107 - 22407834 ----AAUC--AUUCUUUUUCCAGAUCCCCUUGUGUCGCCGAAUUUGGACAUCUCCCGGAUCGGCGCCCGUAUCAUCGAUGAACUGGUGACGGUCAUGAAGGAUGUGGGUAUCG ----....--........((((.((((..(..((.((((((.(((((.......))))))))))).(((..((((((......))))))))).))..).)))).))))..... ( -33.60) >DroEre_CAF1 1746 107 - 1 ----AAUA--AUUUUCCUCGCAGAUCCUCUUGUGUCGCCCAAUUUGGACAUCUCGCGGAUCGGCGCCCGCAUCAUCGAUGAACUGGUGACGGUCAUGAAGGAUGUGGGCAUCG ----....--.........((.(((((.(..(((((..........)))))...).))))).))(((((((((.((.((((.(((....)))))))))..))))))))).... ( -41.50) >DroWil_CAF1 1199 103 - 1 ----U------CUUGUCCUCCAGAUCCACUUGUCUCGCCCAAUUUGGAUAUAUCGAGAAUAGGGGCGCGCAUCAUUGAUGAACUGGUUGUGGUGAUGAAAGAUGUGGGCAUUG ----(------(((((.(((...(((((.(((.......)))..))))).....))).))))))((.((((((.((.((..(((......))).)).)).)))))).)).... ( -25.60) >DroYak_CAF1 1344 107 - 1 ----AACA--AUUUUCGCUGCAGAUCCUCUUGUGUCACCAAAUUUGGACAUCUCGCGGAUCGGCGCCCGCAUUAUCGAUGAACUGGUGACGGUUAUGAAGGAUGUGGGCAUCG ----....--.........((.(((((.(..(((((..........)))))...).))))).))(((((((((.(((...(((((....))))).)))..))))))))).... ( -38.40) >DroMoj_CAF1 1726 107 - 1 ----CUCU--GUCUGUAUUAUAGAUCCCCAUGUCUCGCCCAAUUUGGAUAUAUCACGCAUUGGCGCGCGCAUCAUUGAUGAACUGGUGGUCGUUAUGAAGGACGUGGGCAUCG ----....--............(((.(((((((((((((((((.((((....)).)).))))).)))....((((.(((((.(....).))))))))).))))))))).))). ( -32.50) >DroAna_CAF1 1735 113 - 1 AAAAAAGCAUAUUUGCCUUUCAGAUCCUCUUGUGUCACCCAAUUUGGAUAUCUCGAGGAUCGGAGCCCGCAUCAUUGAUGAGUUAGUGGUUGUGAUGAAGGAUGUGGGCAUCG ......(((....)))...((.(((((((..(((((..........)))))...))))))).))(((((((((..(.(..(.(....).)..).).....))))))))).... ( -33.60) >consensus ____AACA__AUUUGCCUUCCAGAUCCUCUUGUGUCGCCCAAUUUGGACAUCUCGCGGAUCGGCGCCCGCAUCAUCGAUGAACUGGUGACGGUCAUGAAGGAUGUGGGCAUCG ...................((.(((((....(((((..........))))).....))))).))(((((((((....((((.(((....)))))))....))))))))).... (-21.96 = -22.63 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:00 2006