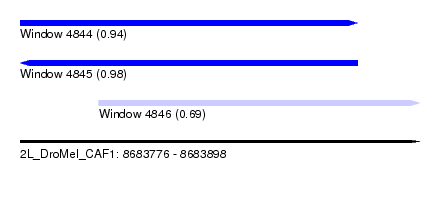
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,683,776 – 8,683,898 |
| Length | 122 |
| Max. P | 0.978281 |

| Location | 8,683,776 – 8,683,879 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 86.30 |
| Mean single sequence MFE | -29.60 |
| Consensus MFE | -27.38 |
| Energy contribution | -27.38 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
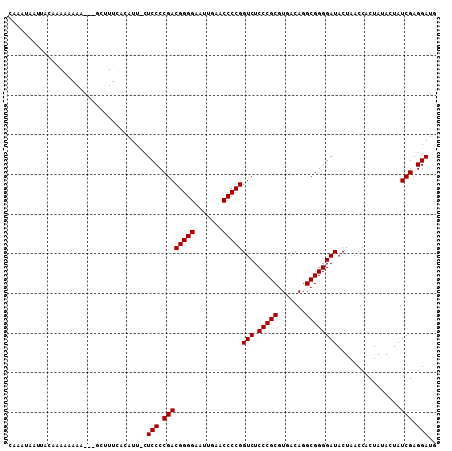
>2L_DroMel_CAF1 8683776 103 + 22407834 CAAAUACUUA---AAAAAA-AUGCUUUCGCAUU-CUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGACG ..........---.....(-((((....)))))-.(((.((((((((.......)))))(((.(((((.......))))))))................))).))).. ( -30.50) >DroPse_CAF1 3309 101 + 1 AUAA-AAUA-UAAAAAGAA-----AUCUACAUUCCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGAUU ....-....-.........-----........(((((.....(((((.......)))))(((.(((((.......))))))))..................))))).. ( -28.30) >DroSec_CAF1 3400 107 + 1 CAUAUAUGUACGAAAAAAAUGUGCUUUCACAUU-CUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGAUG .......(((((.......))))).....((((-(((.....(((((.......)))))(((.(((((.......))))))))..................))))))) ( -31.00) >DroSim_CAF1 2342 106 + 1 CAUAUAUGUACGAAAAAAA-ACGCUUUCACAUU-CUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGAUG ...........((((....-....)))).((((-(((.....(((((.......)))))(((.(((((.......))))))))..................))))))) ( -29.90) >DroPer_CAF1 2243 102 + 1 AUAAAAAUA-UAAAAAGAA-----GUUUACAUUCCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGAUU .........-.........-----........(((((.....(((((.......)))))(((.(((((.......))))))))..................))))).. ( -28.30) >consensus CAAAUAAUUACAAAAAAAA___GCUUUCACAUU_CUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGAUG ...................................(((.((((((((.......)))))(((.(((((.......))))))))................))).))).. (-27.38 = -27.38 + 0.00)

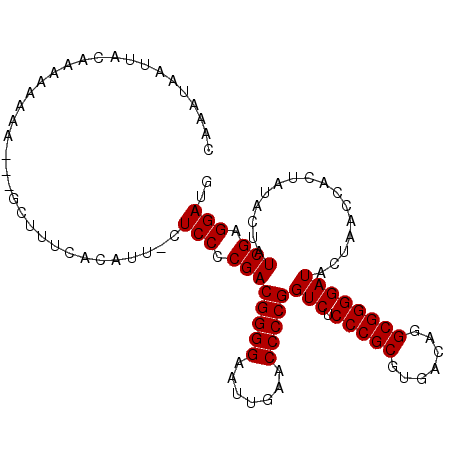
| Location | 8,683,776 – 8,683,879 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 86.30 |
| Mean single sequence MFE | -35.62 |
| Consensus MFE | -29.98 |
| Energy contribution | -29.98 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8683776 103 - 22407834 CGUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAG-AAUGCGAAAGCAU-UUUUUU---UAAGUAUUUG ..((((((((........((((......(((((.......))))).))))((((.......))))))))))))(-(((((....))))-))....---.......... ( -38.20) >DroPse_CAF1 3309 101 - 1 AAUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGGAAUGUAGAU-----UUCUUUUUA-UAUU-UUAU ..((((((((........((((......(((((.......))))).))))((((.......))))))))))))((((((((((.-----.....))))-))))-)).. ( -35.60) >DroSec_CAF1 3400 107 - 1 CAUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAG-AAUGUGAAAGCACAUUUUUUUCGUACAUAUAUG .......(((((....((((.........)))))))))....((....))((((.......))))((((..(((-((((((....)))))))))..)).))....... ( -35.70) >DroSim_CAF1 2342 106 - 1 CAUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAG-AAUGUGAAAGCGU-UUUUUUUCGUACAUAUAUG ..((((((((........((((......(((((.......))))).))))((((.......))))))))))))(-(((((....))))-))................. ( -34.60) >DroPer_CAF1 2243 102 - 1 AAUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGGAAUGUAAAC-----UUCUUUUUA-UAUUUUUAU ..((((((((........((((......(((((.......))))).))))((((.......))))))))))))((((((((((.-----.....))))-))))))... ( -34.00) >consensus CAUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAG_AAUGUGAAAGC___UUUUUUUCAUAAAUAUUUG ..((((((((........((((......(((((.......))))).))))((((.......))))))))))))................................... (-29.98 = -29.98 + 0.00)

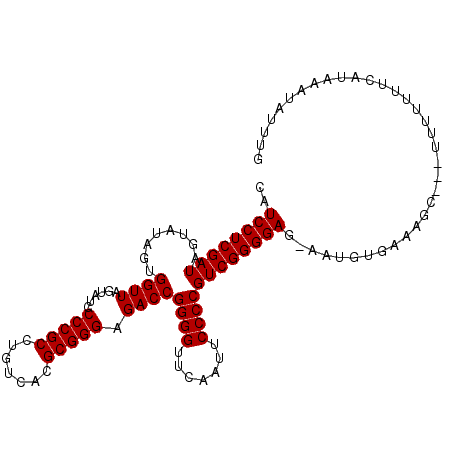
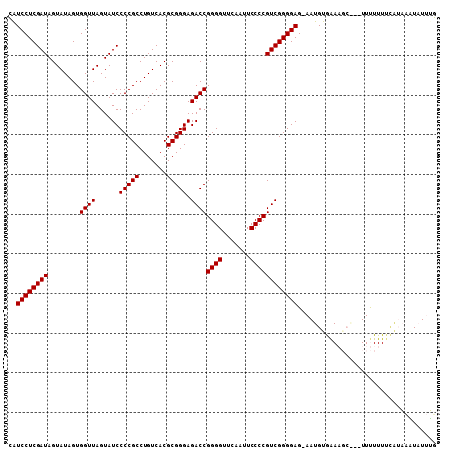
| Location | 8,683,800 – 8,683,898 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.62 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -27.33 |
| Energy contribution | -27.33 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8683800 98 + 22407834 GCAUU-CUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGACGUUGAUACUUUUGAGUCAAA--------------------- .....-.(((.((((((((.......)))))(((.(((((.......))))))))................))).)))..(((((.(....).))))).--------------------- ( -29.90) >DroPse_CAF1 3330 120 + 1 ACAUUCCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGAUUUUCUUCUUCUGCCGUCAAACUGCUAUUUUACCACUAUACU ...........((.((((..((((.....))))..))))))((((.((((((((((.((.......)).))))((((.....)))).))))))))))....................... ( -29.20) >DroEre_CAF1 3581 98 + 1 ACAUU-CUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGACGGUGAACGAGGGAACUCAAA--------------------- .(((.-.(((.((((((((.......)))))(((.(((((.......))))))))................))).)))..)))...(((....)))...--------------------- ( -36.40) >DroYak_CAF1 2381 98 + 1 ACAUU-CUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGACGGUGGACGACCUUAGUCAAA--------------------- .(((.-.(((.((((((((.......)))))(((.(((((.......))))))))................))).)))..)))(((.......)))...--------------------- ( -34.40) >DroAna_CAF1 2137 98 + 1 ACAUU-CUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGACGGUGAUGGACGAGCGCCUAU--------------------- .(((.-.(((.((((((((.......)))))(((.(((((.......))))))))................))).)))..)))..((.(....)))...--------------------- ( -32.10) >DroPer_CAF1 2265 120 + 1 ACAUUCCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGAUUUGCUUCUUCUGCCGUCAAACUGCUAUUUUACCACUAUACU ............(((((((.......))))(((.((((((.......))))))....................(((((......))))).))))))........................ ( -28.80) >consensus ACAUU_CUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGACGGUGAUCGACUGCAGUCAAA_____________________ .......(((.((((((((.......)))))(((.(((((.......))))))))................))).))).......................................... (-27.33 = -27.33 + -0.00)

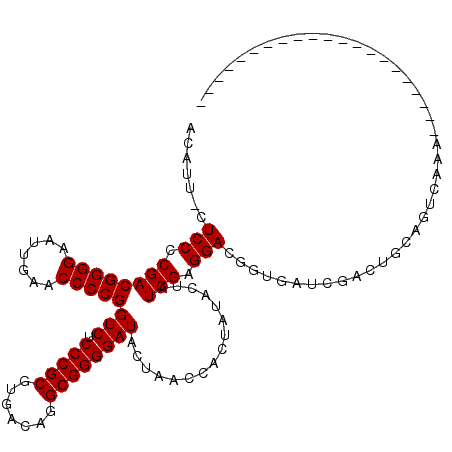
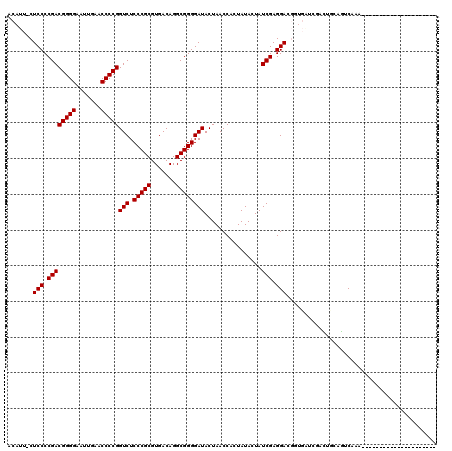
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:58 2006