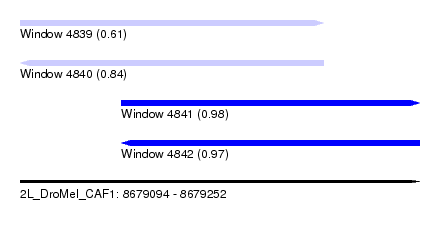
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,679,094 – 8,679,252 |
| Length | 158 |
| Max. P | 0.975248 |

| Location | 8,679,094 – 8,679,214 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.90 |
| Mean single sequence MFE | -37.62 |
| Consensus MFE | -29.93 |
| Energy contribution | -29.93 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8679094 120 + 22407834 UUUUACGAAAUUGAUGCCAAGUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGACGUGAGAGAUUCGUUUAAUUUUUUGU (((((((...(((....))).((((((((........((((......(((((.......))))).))))((((.......))))))))))))..)))))))................... ( -35.50) >DroPse_CAF1 1872 114 + 1 UUUGACGGCGGGAGAACCACAUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAUUCCA---AUUAGUUUUAUUUUU--- ..(((((((((((.((((((((.((....)).)).))))))....).)))))).))))..(((....)))((..((...(((((...)))))))..)).---...............--- ( -40.00) >DroEre_CAF1 2371 114 + 1 UUGUACAAAGGCAGUACCAAGUCCUCGAUAGUUUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGACGCGAGCGAUUCGUAUU--UUU---- ..((((...((.....)).((((((((...(((............(((((.((((.....)))).(((.((((.......))))))))))))))).)))).)))).)))).--...---- ( -35.40) >DroYak_CAF1 2150 113 + 1 UUGUACCAAAGUA-CACCAAGUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAUGCGAGUGAAUCGUAUU--UAU---- .(((((....)))-)).....((((((((........((((......(((((.......))))).))))((((.......))))))))))))(((((((.....)))))))--...---- ( -39.40) >DroAna_CAF1 2694 106 + 1 UUCUACCGAUUGGAUUCCAUGUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGG--------------UUAAUUUUUUUU ..(((.(((..((((.....))))))).)))......((((((..(((((.((((.....)))).(((.((((.......))))))))))))..--------------))))))...... ( -35.40) >DroPer_CAF1 1871 114 + 1 CUUGACGGCGGGAGAACCACAUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAUUCCA---AUUUGUUUUAUUUUU--- ..(((((((((((.((((((((.((....)).)).))))))....).)))))).))))..(((....)))((..((...(((((...)))))))..)).---...............--- ( -40.00) >consensus UUUUACGAAGGGAGUACCAAGUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAUGCGA___AUUCGUUUUAUUUUU___ .....................((((((((........((((......(((((.......))))).))))((((.......))))))))))))............................ (-29.93 = -29.93 + 0.00)

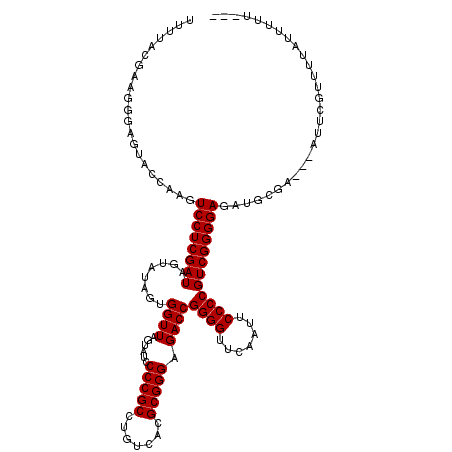
| Location | 8,679,094 – 8,679,214 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.90 |
| Mean single sequence MFE | -34.68 |
| Consensus MFE | -27.33 |
| Energy contribution | -27.33 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8679094 120 - 22407834 ACAAAAAAUUAAACGAAUCUCUCACGUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGACUUGGCAUCAAUUUCGUAAAA ............(((((.......((..(((.((((((((.......)))))(((.(((((.......))))))))................))).)))..))........))))).... ( -33.26) >DroPse_CAF1 1872 114 - 1 ---AAAAAUAAAACUAAU---UGGAAUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGAUGUGGUUCUCCCGCCGUCAAA ---...............---.((.....)).((.((((..((((.....))))..))))))((((.(((((((.....((((((....((....))...)))))).))))))))))).. ( -37.30) >DroEre_CAF1 2371 114 - 1 ----AAA--AAUACGAAUCGCUCGCGUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAAACUAUCGAGGACUUGGUACUGCCUUUGUACAA ----...--....(((.((.((((.((.((((((.(((((.......)))))(((........)))...)))))).))((.....))......)))))).)))((((.......)))).. ( -33.00) >DroYak_CAF1 2150 113 - 1 ----AUA--AAUACGAUUCACUCGCAUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGACUUGGUG-UACUUUGGUACAA ----...--....(((.....)))((..(((.((((((((.......)))))(((.(((((.......))))))))................))).)))..)).((-(((....))))). ( -34.30) >DroAna_CAF1 2694 106 - 1 AAAAAAAAUUAA--------------CCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGACAUGGAAUCCAAUCGGUAGAA ............--------------..((((((.(((((.......)))))(((........)))...))))))..............((((((((((.......)))..))))))).. ( -32.90) >DroPer_CAF1 1871 114 - 1 ---AAAAAUAAAACAAAU---UGGAAUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGAUGUGGUUCUCCCGCCGUCAAG ---...............---.((.....)).((.((((..((((.....))))..))))))((((.(((((((.....((((((....((....))...)))))).))))))))))).. ( -37.30) >consensus ___AAAAAUAAAACGAAU___UCGCAUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGACUUGGUACUCCCUCCGUAAAA ............................(((.((((((((.......)))))(((.(((((.......))))))))................))).)))..................... (-27.33 = -27.33 + -0.00)

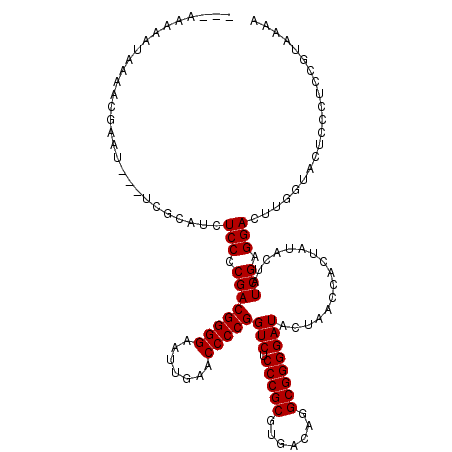
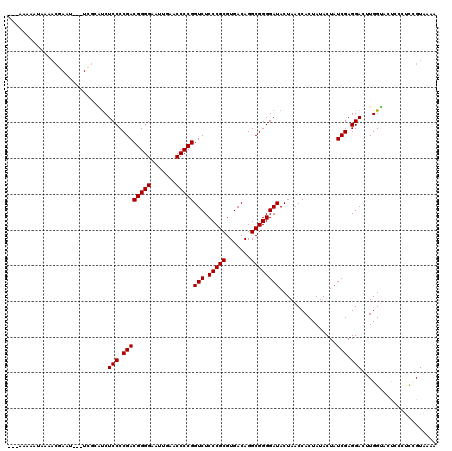
| Location | 8,679,134 – 8,679,252 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.17 |
| Mean single sequence MFE | -35.17 |
| Consensus MFE | -26.79 |
| Energy contribution | -26.57 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975248 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8679134 118 + 22407834 UAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGACGUGAGAGAUUCGUUUAAUUUUUUGU--UGUUCCUCUCUUAGAAUCUUUUUUGUUUUGCUUAUUUA .(((((((((.((((.....)))).(((.((((.......))))))))))))((((.(((((((((..............--.............))))))))).)))).))))...... ( -36.53) >DroSec_CAF1 2367 119 + 1 UAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGACGUGAGAGAUUCGUUUUAUUUUUUGUGUUUUUCCUCUCUUAGAAUCUUU-AUGUUUGGCUUAUUUA .......(((((.......))))).(((.((((.......)))))))((..(((((..((((((.......))))))..)))))..))...............-................ ( -35.50) >DroSim_CAF1 2320 116 + 1 UAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGACGCGAGAGGAUCGUUUUAUUUUUUGUGU---UCCUCUCUUAGAAUCUUU-AUGUUUGGCUUAUUUA ..((.(((((.((((.....)))).(((.((((.......)))))))))))).))(((((((((.((............)).---)))))))...((((....-..)))).))....... ( -38.40) >DroEre_CAF1 2411 103 + 1 UAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGACGCGAGCGAUUCGUAUU--UUU---------UUCUCUCUCAGAA----U-AUG-UUUGCUUAUUUA ..((.(((((.((((.....)))).(((.((((.......)))))))))))).))..((((((..((((((--((.---------.........))))----)-)))-.))))))..... ( -37.00) >DroYak_CAF1 2189 105 + 1 UAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAUGCGAGUGAAUCGUAUU--UAU---------UUGUUUUUCCUCU---UU-UUGUUUUGCUUAUUUA .((((..(((((.......))))).(((.((((.......)))))))(((((((.((((((((((...)))--)))---------)))).))))))).---..-......))))...... ( -36.40) >DroPer_CAF1 1911 91 + 1 UAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAUUCCA---AUUUGUUUUAUUUUU-----------CUUCAGAGAAU--UU-AAUU------------ .....(((((.((((.....)))).(((.((((.......)))))))))))).......---...........((((-----------(....))))).--..-....------------ ( -27.20) >consensus UAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGACGCGAGAGAUUCGUUUUAUUUUU________UCCUCUCUUAGAAU__UU_AUGUUUUGCUUAUUUA ..((.(((((.((((.....)))).(((.((((.......)))))))))))).))................................................................. (-26.79 = -26.57 + -0.22)

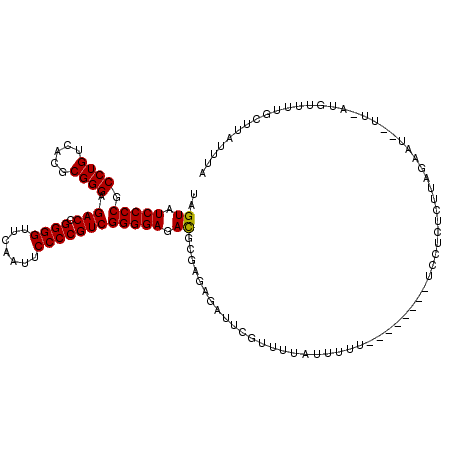
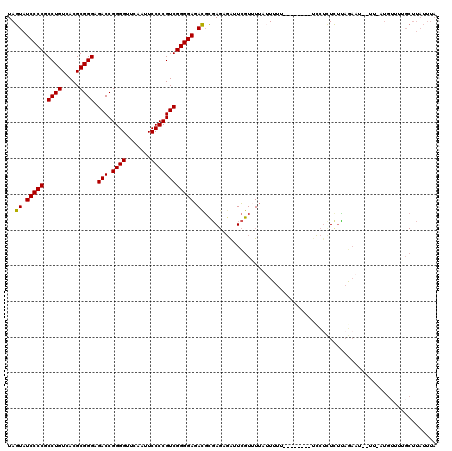
| Location | 8,679,134 – 8,679,252 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.17 |
| Mean single sequence MFE | -29.32 |
| Consensus MFE | -23.67 |
| Energy contribution | -24.00 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8679134 118 - 22407834 UAAAUAAGCAAAACAAAAAAGAUUCUAAGAGAGGAACA--ACAAAAAAUUAAACGAAUCUCUCACGUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUA ............................((((((....--.................))))))..((.((((((.(((((.......)))))(((........)))...)))))).)).. ( -30.30) >DroSec_CAF1 2367 119 - 1 UAAAUAAGCCAAACAU-AAAGAUUCUAAGAGAGGAAAAACACAAAAAAUAAAACGAAUCUCUCACGUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUA ................-...........((((((.......................))))))..((.((((((.(((((.......)))))(((........)))...)))))).)).. ( -30.20) >DroSim_CAF1 2320 116 - 1 UAAAUAAGCCAAACAU-AAAGAUUCUAAGAGAGGA---ACACAAAAAAUAAAACGAUCCUCUCGCGUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUA ................-...........(((((((---.(..............).)))))))..((.((((((.(((((.......)))))(((........)))...)))))).)).. ( -35.54) >DroEre_CAF1 2411 103 - 1 UAAAUAAGCAAA-CAU-A----UUCUGAGAGAGAA---------AAA--AAUACGAAUCGCUCGCGUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUA .......((...-((.-.----...)).(((.((.---------...--........)).)))))((.((((((.(((((.......)))))(((........)))...)))))).)).. ( -29.00) >DroYak_CAF1 2189 105 - 1 UAAAUAAGCAAAACAA-AA---AGAGGAAAAACAA---------AUA--AAUACGAUUCACUCGCAUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUA ................-..---.((((((......---------...--.......))).))).....((((((.(((((.......)))))(((........)))...))))))..... ( -26.09) >DroPer_CAF1 1911 91 - 1 ------------AAUU-AA--AUUCUCUGAAG-----------AAAAAUAAAACAAAU---UGGAAUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUA ------------....-..--.((((....))-----------)).............---(((.((((((((..(((((.......)))))(((........))).)).)))))).))) ( -24.80) >consensus UAAAUAAGCAAAACAU_AA__AUUCUAAGAGAGGA________AAAAAUAAAACGAAUCUCUCGCGUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUA .................................................................((.((((((.(((((.......)))))(((........)))...)))))).)).. (-23.67 = -24.00 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:54 2006