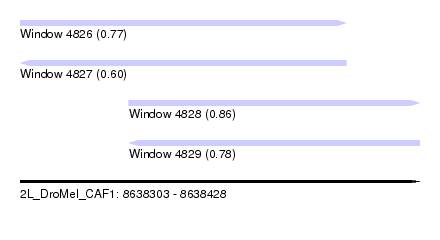
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,638,303 – 8,638,428 |
| Length | 125 |
| Max. P | 0.855476 |

| Location | 8,638,303 – 8,638,405 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 83.97 |
| Mean single sequence MFE | -27.15 |
| Consensus MFE | -22.35 |
| Energy contribution | -23.22 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8638303 102 + 22407834 UAAAUGUGCAACUGCAACAAGGAUGGCAAGA------AGAACAACAACAGGGCCAGGAUAAUACGAGCCAAAGCAGUGGAAAUCCUGGCGUAAUCGUUGCGUUCACUG .....(((.(((.(((((...(((.((....------.......(....).((((((((..(((..((....)).)))...)))))))))).)))))))))))))).. ( -28.40) >DroPse_CAF1 16663 78 + 1 UAAAUGUGCAAUGGCAGCAGC-AUCCU----------------------------ACCAAACACGAG-GAAAGCAGUGAAAAUCCUUGCGUAAUCGUUGCGUUCGCUG .......((((((((....))-.....----------------------------.....((.((((-((............)))))).))...))))))........ ( -15.90) >DroSec_CAF1 7134 102 + 1 UAAAUGUGCAACUGCAACAAGGAUGGCAAGA------AGAACAACAACAGCGCCAGGAUAAUACGAGCCAAAGCAGUGGAAAUCCUGGCGUAAUCGUUGCGUUCACUG .....(((.(((.(((((.....((......------....))......((((((((((..(((..((....)).)))...))))))))))....))))))))))).. ( -31.90) >DroSim_CAF1 7249 102 + 1 UAAAUGUGCAACUGCAACAAGGAUGGCAAGA------AGAACAACAACAGGGCCAGGAUAAUACGAGCCAAAGCAGUGGAAAUCCUGGCGUAAUCGUUGCGUUCACUG .....(((.(((.(((((...(((.((....------.......(....).((((((((..(((..((....)).)))...)))))))))).)))))))))))))).. ( -28.40) >DroEre_CAF1 9735 108 + 1 UAAAUGUGCAACUGCAGCAAGGAUGGCAAGAAGAAGAAGAACAACAACAGGGCCAGGAUAAUACGAGCCAAAGCAGUGGAAAUCCUGGCGUAAUCGUUGCGUUCACUG ......(((....)))((((.(((.((.................(....).((((((((..(((..((....)).)))...)))))))))).))).))))........ ( -28.70) >DroYak_CAF1 7707 102 + 1 UAAAUGUGCAACUGCAACAAGGAUGGCAAGA------GGAACAACAACAUGGCCAGGAUAAUACGAGCCAAAGCAGUGGAAAUCCUGGCGUAAUCGUUGCGUUCACUG .....(((.(((.(((((...(((.((....------(......)......((((((((..(((..((....)).)))...)))))))))).)))))))))))))).. ( -29.60) >consensus UAAAUGUGCAACUGCAACAAGGAUGGCAAGA______AGAACAACAACAGGGCCAGGAUAAUACGAGCCAAAGCAGUGGAAAUCCUGGCGUAAUCGUUGCGUUCACUG .....(((.(((.(((((.................................((((((((..(((..((....)).)))...))))))))(....)))))))))))).. (-22.35 = -23.22 + 0.86)



| Location | 8,638,303 – 8,638,405 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 83.97 |
| Mean single sequence MFE | -29.50 |
| Consensus MFE | -18.44 |
| Energy contribution | -20.97 |
| Covariance contribution | 2.53 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.600871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8638303 102 - 22407834 CAGUGAACGCAACGAUUACGCCAGGAUUUCCACUGCUUUGGCUCGUAUUAUCCUGGCCCUGUUGUUGUUCU------UCUUGCCAUCCUUGUUGCAGUUGCACAUUUA ..((((((((((((((...((((((((.......((....)).......))))))))...))))))))...------...(((((....))..)))))).)))..... ( -30.84) >DroPse_CAF1 16663 78 - 1 CAGCGAACGCAACGAUUACGCAAGGAUUUUCACUGCUUUC-CUCGUGUUUGGU----------------------------AGGAU-GCUGCUGCCAUUGCACAUUUA .((.(((.(((..((...(....).....))..))).)))-)).((((.((((----------------------------((...-....))))))..))))..... ( -19.40) >DroSec_CAF1 7134 102 - 1 CAGUGAACGCAACGAUUACGCCAGGAUUUCCACUGCUUUGGCUCGUAUUAUCCUGGCGCUGUUGUUGUUCU------UCUUGCCAUCCUUGUUGCAGUUGCACAUUUA ..((((((((((((((..(((((((((.......((....)).......)))))))))..))))))))...------...(((((....))..)))))).)))..... ( -34.24) >DroSim_CAF1 7249 102 - 1 CAGUGAACGCAACGAUUACGCCAGGAUUUCCACUGCUUUGGCUCGUAUUAUCCUGGCCCUGUUGUUGUUCU------UCUUGCCAUCCUUGUUGCAGUUGCACAUUUA ..((((((((((((((...((((((((.......((....)).......))))))))...))))))))...------...(((((....))..)))))).)))..... ( -30.84) >DroEre_CAF1 9735 108 - 1 CAGUGAACGCAACGAUUACGCCAGGAUUUCCACUGCUUUGGCUCGUAUUAUCCUGGCCCUGUUGUUGUUCUUCUUCUUCUUGCCAUCCUUGCUGCAGUUGCACAUUUA ..((((((((((((((...((((((((.......((....)).......))))))))...))))))))............(((((....))..)))))).)))..... ( -30.84) >DroYak_CAF1 7707 102 - 1 CAGUGAACGCAACGAUUACGCCAGGAUUUCCACUGCUUUGGCUCGUAUUAUCCUGGCCAUGUUGUUGUUCC------UCUUGCCAUCCUUGUUGCAGUUGCACAUUUA ..((((((((((((((...((((((((.......((....)).......))))))))...))))))))...------...(((((....))..)))))).)))..... ( -30.84) >consensus CAGUGAACGCAACGAUUACGCCAGGAUUUCCACUGCUUUGGCUCGUAUUAUCCUGGCCCUGUUGUUGUUCU______UCUUGCCAUCCUUGUUGCAGUUGCACAUUUA ........((((((((...((((((((.......((....)).......))))))))...)))))))).....................((.(((....))))).... (-18.44 = -20.97 + 2.53)

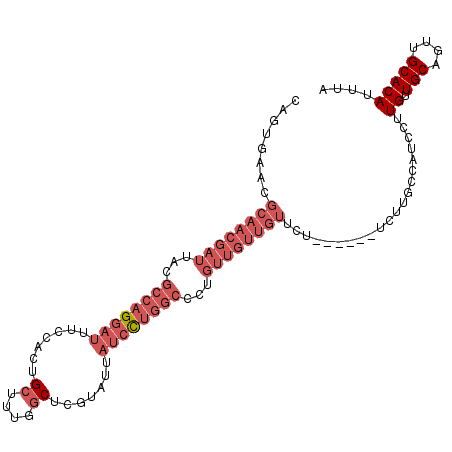

| Location | 8,638,337 – 8,638,428 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 81.35 |
| Mean single sequence MFE | -27.56 |
| Consensus MFE | -17.56 |
| Energy contribution | -17.95 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8638337 91 + 22407834 ACAACAACAGGGCCAGGAUAAUACGAGCCAAAGCAGUGGAAAUCCUGGCGUAAUCGUUGCGUUCACUGCUGGCAGGCAACAACAAC------------AGCAA .((((....(.((((((((..(((..((....)).)))...)))))))).)....)))).......(((((...(....).....)------------)))). ( -27.40) >DroPse_CAF1 16689 76 + 1 ---------------ACCAAACACGAG-GAAAGCAGUGAAAAUCCUUGCGUAAUCGUUGCGUUCGCUGCUGC-GGGGAA-AACUACUGC---------AACAC ---------------.((........)-)...((((((....((((((((.....((.((....)).)))))-))))).-...))))))---------..... ( -21.00) >DroSec_CAF1 7168 94 + 1 ACAACAACAGCGCCAGGAUAAUACGAGCCAAAGCAGUGGAAAUCCUGGCGUAAUCGUUGCGUUCACUGCUGGCAGGCAACAACAACUGG---------AACAA .((((....((((((((((..(((..((....)).)))...))))))))))....)))).((((..((((....)))).((.....)))---------))).. ( -30.70) >DroSim_CAF1 7283 94 + 1 ACAACAACAGGGCCAGGAUAAUACGAGCCAAAGCAGUGGAAAUCCUGGCGUAAUCGUUGCGUUCACUGCUGGCAGGCAACAACAACUGG---------AACAA .........(..((((..........((((..((((((((......((((....))))...)))))))))))).(....).....))))---------..).. ( -28.90) >DroEre_CAF1 9775 103 + 1 ACAACAACAGGGCCAGGAUAAUACGAGCCAAAGCAGUGGAAAUCCUGGCGUAAUCGUUGCGUUCACUGCUGGCAGGCAACAACAACUGCAACAACUGGAACAA .......(((.((((((((..(((..((....)).)))...))))))))......((((((((...((((....)))).....))).)))))..)))...... ( -31.40) >DroYak_CAF1 7741 103 + 1 ACAACAACAUGGCCAGGAUAAUACGAGCCAAAGCAGUGGAAAUCCUGGCGUAAUCGUUGCGUUCACUGCUCGCUGGCAACAACAACUGGAACCACUACAACAA .........(((((((..........((((.(((((((((......((((....))))...)))))))))...))))........))))..)))......... ( -25.97) >consensus ACAACAACAGGGCCAGGAUAAUACGAGCCAAAGCAGUGGAAAUCCUGGCGUAAUCGUUGCGUUCACUGCUGGCAGGCAACAACAACUGG_________AACAA ..........................(((..(((((((((......((((....))))...)))))))))....))).......................... (-17.56 = -17.95 + 0.39)


| Location | 8,638,337 – 8,638,428 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 81.35 |
| Mean single sequence MFE | -30.85 |
| Consensus MFE | -19.09 |
| Energy contribution | -22.12 |
| Covariance contribution | 3.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8638337 91 - 22407834 UUGCU------------GUUGUUGUUGCCUGCCAGCAGUGAACGCAACGAUUACGCCAGGAUUUCCACUGCUUUGGCUCGUAUUAUCCUGGCCCUGUUGUUGU (..((------------((((..((.....))))))))..)..((((((((...((((((((.......((....)).......))))))))...)))))))) ( -33.24) >DroPse_CAF1 16689 76 - 1 GUGUU---------GCAGUAGUU-UUCCCC-GCAGCAGCGAACGCAACGAUUACGCAAGGAUUUUCACUGCUUUC-CUCGUGUUUGGU--------------- .((((---------((.......-......-)))))).(((((((...((....(((..(.....)..)))..))-...)))))))..--------------- ( -15.62) >DroSec_CAF1 7168 94 - 1 UUGUU---------CCAGUUGUUGUUGCCUGCCAGCAGUGAACGCAACGAUUACGCCAGGAUUUCCACUGCUUUGGCUCGUAUUAUCCUGGCGCUGUUGUUGU ..(((---------(...((((((........)))))).))))((((((((..(((((((((.......((....)).......)))))))))..)))))))) ( -35.24) >DroSim_CAF1 7283 94 - 1 UUGUU---------CCAGUUGUUGUUGCCUGCCAGCAGUGAACGCAACGAUUACGCCAGGAUUUCCACUGCUUUGGCUCGUAUUAUCCUGGCCCUGUUGUUGU ..(((---------(...((((((........)))))).))))((((((((...((((((((.......((....)).......))))))))...)))))))) ( -31.84) >DroEre_CAF1 9775 103 - 1 UUGUUCCAGUUGUUGCAGUUGUUGUUGCCUGCCAGCAGUGAACGCAACGAUUACGCCAGGAUUUCCACUGCUUUGGCUCGUAUUAUCCUGGCCCUGUUGUUGU ..(((((.((((..((((..(....)..)))))))).).))))((((((((...((((((((.......((....)).......))))))))...)))))))) ( -35.44) >DroYak_CAF1 7741 103 - 1 UUGUUGUAGUGGUUCCAGUUGUUGUUGCCAGCGAGCAGUGAACGCAACGAUUACGCCAGGAUUUCCACUGCUUUGGCUCGUAUUAUCCUGGCCAUGUUGUUGU (..((((.(((((..(((...)))..)))).)..))))..)..((((((((...((((((((.......((....)).......))))))))...)))))))) ( -33.74) >consensus UUGUU_________CCAGUUGUUGUUGCCUGCCAGCAGUGAACGCAACGAUUACGCCAGGAUUUCCACUGCUUUGGCUCGUAUUAUCCUGGCCCUGUUGUUGU ..................(..((((((.....))))))..)..((((((((...((((((((.......((....)).......))))))))...)))))))) (-19.09 = -22.12 + 3.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:42 2006