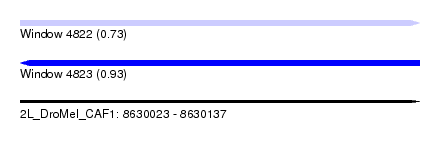
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,630,023 – 8,630,137 |
| Length | 114 |
| Max. P | 0.930860 |

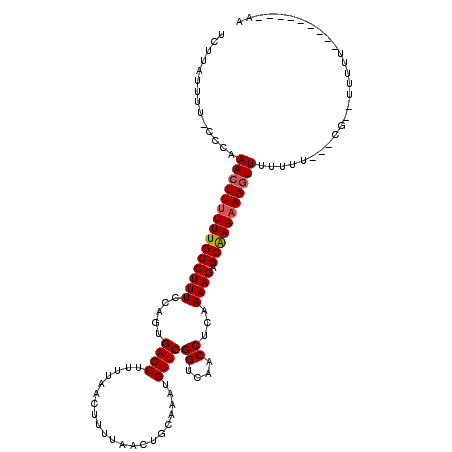
| Location | 8,630,023 – 8,630,137 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 83.17 |
| Mean single sequence MFE | -19.20 |
| Consensus MFE | -18.46 |
| Energy contribution | -18.68 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8630023 114 + 22407834 UUGAAAUACAAAAAAAAACG---AGAAAAACUUUUUUGCUCUUUUGAGGUUGACCGAACAUUUGCAGUUAAAAGUUAAAAGCUCACUGGAAAGGCAAAAAAGUUGGGG-AAAAUAAGA ....................---.....(((((((((((.(((((.((.(((((.(........).))))).(((.....)))..)).))))))))))))))))....-......... ( -20.80) >DroSec_CAF1 58230 99 + 1 UUGA-------A-----------AAAAAAACUUGUUUGCUCUUUUGGGGUUGACCGAACAUUUGCAGUUAAAAGUUAAAAGCUCACUGGAAAGGCAAAAAAGUUUGGG-AAAAUAAGA ....-------.-----------.......(((((((((((.....))))...((((((.(((((.......(((.....)))..((....)))))))...)))))).-.))))))). ( -19.40) >DroSim_CAF1 64534 101 + 1 UUGGAA-----A-----------AAAAAAACUUUUUUGCUCUUUUGAGGUUGACCGAACAUUUGCAGUUAAAAGUUAAAAGCUCACUGGAAAGGCAAAAAAGUUUGGG-AAAAUAAGA ......-----.-----------....((((((((((((.(((((.((.(((((.(........).))))).(((.....)))..)).)))))))))))))))))...-......... ( -21.70) >DroEre_CAF1 59578 105 + 1 AA---------AAAAAAGCGAG---AACAACUUUUUUGCUCUUUUGAGGUUGACCGAACAUUUGCAGUUAAAAGUUAAAAGCUCACUGGAAAGGCAAAAAAUUUUGGA-AAAAUAAGA ..---------((((.((((((---((.....)))))))).))))........(((((..(((((.......(((.....)))..((....)))))))....))))).-......... ( -16.30) >DroYak_CAF1 61214 108 + 1 CA---------AAAAACGAGAAAAAAACAACUUUUUUGCUCUUUUGAGGUUGACCGAACAUUUGCAGUUAAAAGUUAAAAGCUCACUGGAAAGGCAAAAAAGUUUGGG-AAAAUAAGA ..---------................((((((((((((.(((((.((.(((((.(........).))))).(((.....)))..)).)))))))))))))).)))..-......... ( -20.40) >DroAna_CAF1 61456 104 + 1 GA---------AAAAA--CG---AAGAGAACUUUUUCGCUCUUUUGUGGUUGACCGAACAUUUGCAGUUAAAAGUUAAAAGCUCGCUGAAAAGGCAAAAAAGUUGAGGAAAAAUGGGG ..---------.....--..---.....((((((((.((..((((((((((.....(((.((((....)))).)))...))).))).))))..)).)))))))).............. ( -16.60) >consensus UA_________AAAAA__CG___AAAAAAACUUUUUUGCUCUUUUGAGGUUGACCGAACAUUUGCAGUUAAAAGUUAAAAGCUCACUGGAAAGGCAAAAAAGUUUGGG_AAAAUAAGA ............................(((((((((((.(((((..((....)).........((((....(((.....))).)))))))))))))))))))).............. (-18.46 = -18.68 + 0.22)

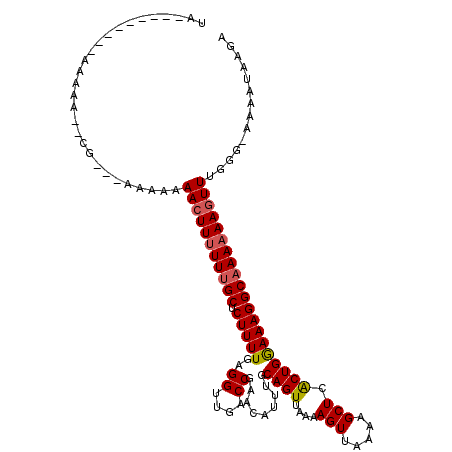
| Location | 8,630,023 – 8,630,137 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 83.17 |
| Mean single sequence MFE | -16.48 |
| Consensus MFE | -15.81 |
| Energy contribution | -16.00 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8630023 114 - 22407834 UCUUAUUUU-CCCCAACUUUUUUGCCUUUCCAGUGAGCUUUUAACUUUUAACUGCAAAUGUUCGGUCAACCUCAAAAGAGCAAAAAAGUUUUUCU---CGUUUUUUUUUGUAUUUCAA .........-....(((((((((((((((.....((((.....................))))((....))...)))).))))))))))).....---.................... ( -16.80) >DroSec_CAF1 58230 99 - 1 UCUUAUUUU-CCCAAACUUUUUUGCCUUUCCAGUGAGCUUUUAACUUUUAACUGCAAAUGUUCGGUCAACCCCAAAAGAGCAAACAAGUUUUUUU-----------U-------UCAA .........-...((((((.(((((((((.....((((.....................))))((......)).)))).))))).))))))....-----------.-------.... ( -14.00) >DroSim_CAF1 64534 101 - 1 UCUUAUUUU-CCCAAACUUUUUUGCCUUUCCAGUGAGCUUUUAACUUUUAACUGCAAAUGUUCGGUCAACCUCAAAAGAGCAAAAAAGUUUUUUU-----------U-----UUCCAA .........-...((((((((((((((((.....((((.....................))))((....))...)))).))))))))))))....-----------.-----...... ( -17.70) >DroEre_CAF1 59578 105 - 1 UCUUAUUUU-UCCAAAAUUUUUUGCCUUUCCAGUGAGCUUUUAACUUUUAACUGCAAAUGUUCGGUCAACCUCAAAAGAGCAAAAAAGUUGUU---CUCGCUUUUUU---------UU .........-.....................((((((....((((((((...(((........((....))((....))))).))))))))..---)))))).....---------.. ( -14.80) >DroYak_CAF1 61214 108 - 1 UCUUAUUUU-CCCAAACUUUUUUGCCUUUCCAGUGAGCUUUUAACUUUUAACUGCAAAUGUUCGGUCAACCUCAAAAGAGCAAAAAAGUUGUUUUUUUCUCGUUUUU---------UG .........-....(((((((((((((((.....((((.....................))))((....))...)))).))))))))))).................---------.. ( -17.40) >DroAna_CAF1 61456 104 - 1 CCCCAUUUUUCCUCAACUUUUUUGCCUUUUCAGCGAGCUUUUAACUUUUAACUGCAAAUGUUCGGUCAACCACAAAAGAGCGAAAAAGUUCUCUU---CG--UUUUU---------UC ..............((((((((((((((((....((((.....................))))((....))..))))).))))))))))).....---..--.....---------.. ( -18.20) >consensus UCUUAUUUU_CCCAAACUUUUUUGCCUUUCCAGUGAGCUUUUAACUUUUAACUGCAAAUGUUCGGUCAACCUCAAAAGAGCAAAAAAGUUUUUUU___CG__UUUUU_________AA ..............(((((((((((((((.....((((.....................))))((....))...)))).)))))))))))............................ (-15.81 = -16.00 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:37 2006