
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 872,785 – 872,887 |
| Length | 102 |
| Max. P | 0.933296 |

| Location | 872,785 – 872,887 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.37 |
| Mean single sequence MFE | -23.02 |
| Consensus MFE | -12.27 |
| Energy contribution | -13.38 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
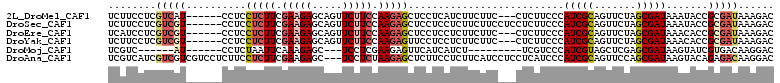
>2L_DroMel_CAF1 872785 102 + 22407834 UCUUCCUCGUCAU------CCUCCUCUUCGAAGAGCAGUUCUUCCAAGAGCUCCUCAUCUUCUUC---CUCUUCCCAUCGCAGUUCUAGCGAUAAAUACCGCGAUAAAGAC ((((..((((...------..........((.(((.((((((....)))))).))).))......---........(((((.......))))).......))))..)))). ( -22.00) >DroSec_CAF1 1158 105 + 1 UCUUCCUCGUCGU------CCUCCUCUUCGAAGAGCAGUUCUUCCAAGAGCUCCUCCUCUUCUUCCUCCUCUUCCCAUCGCAGUUCUAGCGAUAAAUACCGCGAUAAAGAC ((((..((((.((------..........((((((.((((((....))))))....))))))..............(((((.......)))))....)).))))..)))). ( -24.20) >DroEre_CAF1 4415 102 + 1 UCAUCCUCGUCGU------CCUCCUCUUCGAAGAGCAGUUCUUCCAAGAGCUCCUCCUCUUCUUC---CUCUUCCCAUCGCAGUUCUAGCGAUAAACACCGCGAUAAAGAC ........(((((------..........((((((.((((((....))))))....))))))...---........(((((.......))))).......)))))...... ( -21.70) >DroYak_CAF1 919 102 + 1 UCUUCCUCGUCGU------CCUCCUCUUCGAAGAGCAGUUCUUCCAAGAGUUCCUCCUCUUCUUC---CUCUUCCCAUCGCAGUUCUAGCGAUAAACACCGCGAUAAAGAC ((((..((((.((------....(((((.((((((...)))))).)))))...............---........(((((.......)))))....)).))))..)))). ( -22.00) >DroMoj_CAF1 1100 87 + 1 UCGUC------AU------CCUCUAAUUCAAAGAGC---UCCUCGAAGAGUUCAUCAUCU---------UCGUCCCAUCGUAGCUCGAGCGAUAAGUAUCGUGACAAGGAC ..(((------..------.............((((---((..((((((........)))---------))).......).)))))..(((((....))))))))...... ( -19.50) >DroAna_CAF1 1137 108 + 1 UCGUCAUCGUCGUCGUCCUCUUCCUCUUCGAAGAGC---UCCUCUAAGAGCUCUUCCUCUUCAUCCUCCUCAUCCCAUCGCAGUUCCAGCGAUAAGUACAGAGACAAGGAC ..............(((((..((.(((..(((((((---((......)))))))))....................(((((.......)))))......)))))..))))) ( -28.70) >consensus UCUUCCUCGUCGU______CCUCCUCUUCGAAGAGCAGUUCUUCCAAGAGCUCCUCCUCUUCUUC___CUCUUCCCAUCGCAGUUCUAGCGAUAAAUACCGCGAUAAAGAC ........(((((..........(((((.(((((.....))))).)))))..........................(((((.......))))).......)))))...... (-12.27 = -13.38 + 1.11)
| Location | 872,785 – 872,887 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.37 |
| Mean single sequence MFE | -30.18 |
| Consensus MFE | -20.36 |
| Energy contribution | -21.17 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
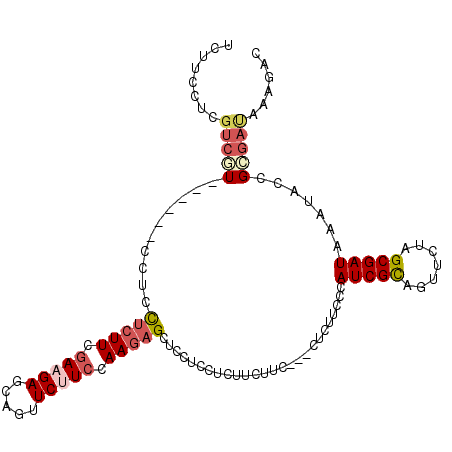
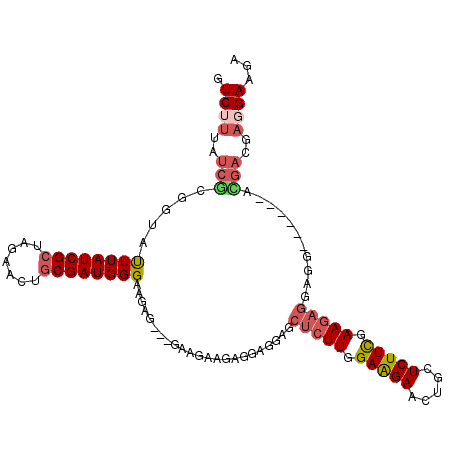
>2L_DroMel_CAF1 872785 102 - 22407834 GUCUUUAUCGCGGUAUUUAUCGCUAGAACUGCGAUGGGAAGAG---GAAGAAGAUGAGGAGCUCUUGGAAGAACUGCUCUUCGAAGAGGAGG------AUGACGAGGAAGA ((((((((((((((.((((....))))))))))))))).....---...............(((((.(((((.....))))).)))))....------..)))........ ( -28.00) >DroSec_CAF1 1158 105 - 1 GUCUUUAUCGCGGUAUUUAUCGCUAGAACUGCGAUGGGAAGAGGAGGAAGAAGAGGAGGAGCUCUUGGAAGAACUGCUCUUCGAAGAGGAGG------ACGACGAGGAAGA ((((((((((((((.((((....))))))))))))..((((((.((....(((((......))))).......)).))))))......))))------))........... ( -31.20) >DroEre_CAF1 4415 102 - 1 GUCUUUAUCGCGGUGUUUAUCGCUAGAACUGCGAUGGGAAGAG---GAAGAAGAGGAGGAGCUCUUGGAAGAACUGCUCUUCGAAGAGGAGG------ACGACGAGGAUGA ((((((((((((((.((((....))))))))))))........---......((((((.((.(((....))).)).))))))......))))------))........... ( -29.40) >DroYak_CAF1 919 102 - 1 GUCUUUAUCGCGGUGUUUAUCGCUAGAACUGCGAUGGGAAGAG---GAAGAAGAGGAGGAACUCUUGGAAGAACUGCUCUUCGAAGAGGAGG------ACGACGAGGAAGA ((((((((((((((.((((....))))))))))))........---...............(((((.(((((.....))))).)))))))))------))........... ( -29.70) >DroMoj_CAF1 1100 87 - 1 GUCCUUGUCACGAUACUUAUCGCUCGAGCUACGAUGGGACGA---------AGAUGAUGAACUCUUCGAGGA---GCUCUUUGAAUUAGAGG------AU------GACGA ((((((.(((((((....))))...(((((.(....)..(((---------(((........))))))...)---))))..)))....))))------))------..... ( -21.80) >DroAna_CAF1 1137 108 - 1 GUCCUUGUCUCUGUACUUAUCGCUGGAACUGCGAUGGGAUGAGGAGGAUGAAGAGGAAGAGCUCUUAGAGGA---GCUCUUCGAAGAGGAAGAGGACGACGACGAUGACGA ((((((.(((((...((((((((.......)))))))).................((((((((((....)))---)))))))..)))))..)))))).............. ( -41.00) >consensus GUCUUUAUCGCGGUAUUUAUCGCUAGAACUGCGAUGGGAAGAG___GAAGAAGAGGAGGAGCUCUUGGAAGAACUGCUCUUCGAAGAGGAGG______ACGACGAGGAAGA .((((..(((.....((((((((.......)))))))).......................(((((.(((((.....))))).)))))...........)))..))))... (-20.36 = -21.17 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:31 2006