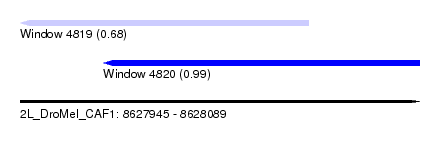
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,627,945 – 8,628,089 |
| Length | 144 |
| Max. P | 0.994867 |

| Location | 8,627,945 – 8,628,049 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 74.44 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -7.66 |
| Energy contribution | -7.86 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.29 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
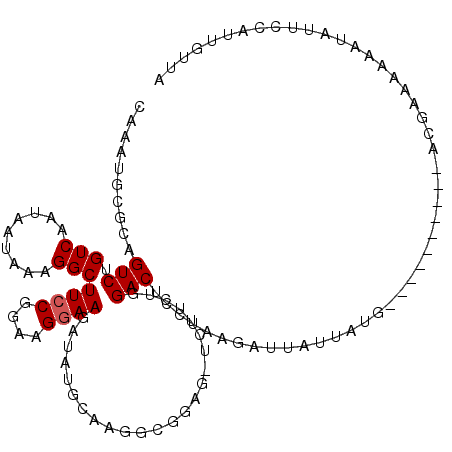
>2L_DroMel_CAF1 8627945 104 - 22407834 CAAAU--GCAGUCUGUCAAUAAUAAAGGCUUCCGGAAGGAAGUUACACAAUGCGGAGGUCUUCUGGACUCCUAAGUUUAUUAUG----------AGGAAAAAAUAUUCCAUUGUUA .....--(((((...(((.((((((((((((((....))))))).........((((..(....)..))))....)))))))))----------)((((......))))))))).. ( -28.70) >DroSec_CAF1 56160 95 - 1 CAAAUGUGCAGUCAGUCAAUAAUAAAGGCUUCCGGAAGGAAGAUAUGCAAGGCG-----------GACUCCUAAGAUUAUAAAG----------ACGAAAAAAUAUUCCAUUGUUA ..(((((....((.(((.(((((..((((((((....)))))...(((...)))-----------....)))...)))))...)----------))))....)))))......... ( -17.40) >DroSim_CAF1 62515 95 - 1 CAAAUGUGCAGUCUGUCAAUAAUAAAGGCUUCCGGAAGGAAGAUAUGCAAGGCG-----------GACUCCUAAGAUUAUAAAG----------ACGAAAAAAUAUUCCAUUGUUA .......(.((((((((....(((....(((((....))))).)))....))))-----------)))).)............(----------((((............))))). ( -20.50) >DroEre_CAF1 57419 116 - 1 CAACGGCAAAGUCUGUCAAUAAUGAAGGCUUCCGGAAGAAAAAUAUGCAAGGCAGAGUUCUUCUGGACUCCUAAGAUGAUUAUGAACGGAGCAGACGAAAAAACGUGCGAUUGUUG ...((((((..(((((..(((((.((((..((((((((((.....(((...)))...))))))))))..)))....).)))))..)))))((..(((......)))))..)))))) ( -32.70) >DroYak_CAF1 59078 114 - 1 CAACGGCA--GUCUGUCAAUAAUGAAGGCUUCCGGUAGGAAAAUAUGCAAGGCGGAGGUCUUCUGGACUCCUAAGAUGAUUAUGAACAGAUCAUGCGAAAAAACGUUCGUUUCUGG ....((.(--(((((........(((((((((((.(..(........)..).)))))))))))))))))))...............((((....(((((......))))).)))). ( -30.80) >consensus CAAAUGCGCAGUCUGUCAAUAAUAAAGGCUUCCGGAAGGAAGAUAUGCAAGGCGGAG_UCUUCUGGACUCCUAAGAUUAUUAUG__________ACGAAAAAAUAUUCCAUUGUUA ..........(((.(((.........)))((((....))))........................)))................................................ ( -7.66 = -7.86 + 0.20)

| Location | 8,627,975 – 8,628,089 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 75.54 |
| Mean single sequence MFE | -32.06 |
| Consensus MFE | -12.88 |
| Energy contribution | -14.20 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.40 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
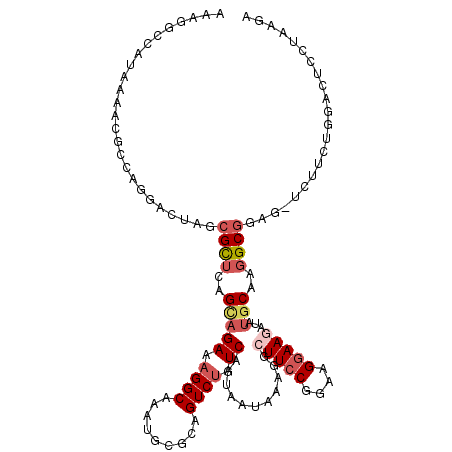
>2L_DroMel_CAF1 8627975 114 - 22407834 AAUGGCCAUAAAACGCCGGGACUAGCGCUCAGCAGAAAGGCAAAU--GCAGUCUGUCAAUAAUAAAGGCUUCCGGAAGGAAGUUACACAAUGCGGAGGUCUUCUGGACUCCUAAGU ...(((........)))((((((..(((...(((((...((....--))..)))))..........(((((((....))))))).......))).))(((.....))))))).... ( -33.80) >DroSec_CAF1 56190 105 - 1 AAAGGCCAUAAAACGCCAGGACUAGCGCUCAGCAGAAAGGCAAAUGUGCAGUCAGUCAAUAAUAAAGGCUUCCGGAAGGAAGAUAUGCAAGGCG-----------GACUCCUAAGA ...(((........)))((((.(..((((..(((((...(((....)))..))...............(((((....)))))...)))..))))-----------.).)))).... ( -30.00) >DroSim_CAF1 62545 105 - 1 AAAGGCCAUAAAACGGCAGGACUAGCGCUCAGCAGAAAGGCAAAUGUGCAGUCUGUCAAUAAUAAAGGCUUCCGGAAGGAAGAUAUGCAAGGCG-----------GACUCCUAAGA ....(((.......)))((((...((((...((......))....)))).(((((((....(((....(((((....))))).)))....))))-----------))))))).... ( -33.80) >DroEre_CAF1 57459 116 - 1 AAAGGAUACAAAACGCCAGGACUAGAGCUCCGCAGAAAGGCAACGGCAAAGUCUGUCAAUAAUGAAGGCUUCCGGAAGAAAAAUAUGCAAGGCAGAGUUCUUCUGGACUCCUAAGA ..((((........(((.((((....).)))(((((...((....))....)))))..........))).((((((((((.....(((...)))...)))))))))).)))).... ( -33.90) >DroYak_CAF1 59118 93 - 1 AAGG---------------------CGUUCCGUAGAAAGGCAACGGCA--GUCUGUCAAUAAUGAAGGCUUCCGGUAGGAAAAUAUGCAAGGCGGAGGUCUUCUGGACUCCUAAGA .(((---------------------.((((.(((((...((....)).--.)))).)......(((((((((((.(..(........)..).))))))))))).)))).))).... ( -28.80) >consensus AAAGGCCAUAAAACGCCAGGACUAGCGCUCAGCAGAAAGGCAAAUGCGCAGUCUGUCAAUAAUAAAGGCUUCCGGAAGGAAGAUAUGCAAGGCGGAG_UCUUCUGGACUCCUAAGA .........................((((..(((((.((((.........)))).))...........(((((....)))))...)))..))))...................... (-12.88 = -14.20 + 1.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:34 2006