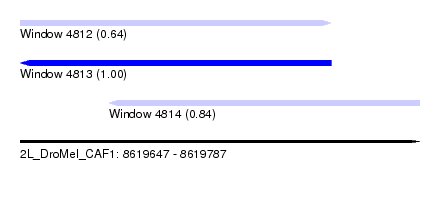
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,619,647 – 8,619,787 |
| Length | 140 |
| Max. P | 0.997659 |

| Location | 8,619,647 – 8,619,756 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 71.72 |
| Mean single sequence MFE | -29.22 |
| Consensus MFE | -14.02 |
| Energy contribution | -15.42 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8619647 109 + 22407834 UUGCUGGUCCCUGCAGCUU---GUAGCUCAGC--UUGGCUCC-CCGGUUGAAAGCCCACCAGAAGCAGCAUCAGCAGCAGCACCAGCUUGUAU-UUCAUUGUUUUAAAACAAUGUU ..((((((..((((.(((.---((.(((..((--((((....-))(((.(.....).)))..))))))))).))).)))).))))))......-..(((((((....))))))).. ( -32.70) >DroVir_CAF1 63188 93 + 1 UCGUUGGUU-----AGCUUUCGGGCGCUCAGU--UGGGCUCC---AGCUCUGAGCCCUGGG-CU----CCUC--------UUUUACCAUGUAUUUUCAUUGUUUUAGAACAAUGUU ....((((.-----.......(((.(((((..--.((((((.---......))))))))))-).----))).--------....))))........(((((((....))))))).. ( -28.02) >DroSec_CAF1 47903 109 + 1 UUGCUGGUCCUUGCAGCUU---GUAGCUCAGC--UUGGCUCC-CCGGUUGAAAGCCCACCAGAAGCAGCAGCAGCGGCAGCACCAGCUUGUAU-UUCAUUGUUUUAAAACAAUGUU ..((((((.((.((.(((.---((.(((..((--((((....-))(((.(.....).)))..))))))).))))).)))).))))))......-..(((((((....))))))).. ( -33.50) >DroSim_CAF1 53553 109 + 1 UUGCUGGUCCCUGCAGCUU---GUAGCUCAGC--UUGGCUCC-CCGGUUGAAAGCCCACCAGAAGCAGCAGCAGCGGCAGCACCAGCUUGUAU-UUCAUUGUUUUAAAACAAUGUU ..((((((..((((.(((.---((.(((..((--((((....-))(((.(.....).)))..))))))).))))).)))).))))))......-..(((((((....))))))).. ( -35.10) >DroYak_CAF1 50876 93 + 1 UUGCUGGUCCCUGCAGCUU---GUAGCUCAGC--UCGGCUCCCCCGGUUGAAAGCCCACCAGC-----------------CAGCAGCUUGUAU-UUCAUUGUUUUAAAACAAUGUU ((((((((..((((.....---)))).(((((--.(((.....))))))))..........))-----------------)))))).......-..(((((((....))))))).. ( -26.90) >DroAna_CAF1 50926 94 + 1 UUGUUGGUCUCUGC--CAU---GUAGCUCAGUCGUUGCCCCU-CCGC------GCCCCCUC-UU---GCUUC-----UAUGACCGGCUUGUAU-UUCAUUGUUUUAAAACAAUGUU ..(((((((.....--...---(((((......)))))....-..((------(.......-.)---))...-----...)))))))......-..(((((((....))))))).. ( -19.10) >consensus UUGCUGGUCCCUGCAGCUU___GUAGCUCAGC__UUGGCUCC_CCGGUUGAAAGCCCACCAGAA___GCAUC_____CAGCACCAGCUUGUAU_UUCAUUGUUUUAAAACAAUGUU ..((((((..(((.((((......))))))).....((((............)))).........................)))))).........(((((((....))))))).. (-14.02 = -15.42 + 1.39)

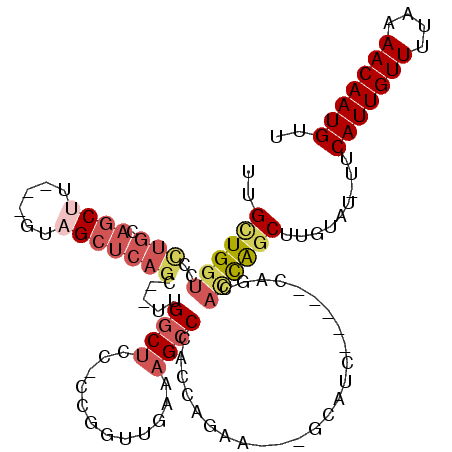
| Location | 8,619,647 – 8,619,756 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 71.72 |
| Mean single sequence MFE | -34.18 |
| Consensus MFE | -16.79 |
| Energy contribution | -19.68 |
| Covariance contribution | 2.89 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.49 |
| SVM decision value | 2.90 |
| SVM RNA-class probability | 0.997659 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8619647 109 - 22407834 AACAUUGUUUUAAAACAAUGAA-AUACAAGCUGGUGCUGCUGCUGAUGCUGCUUCUGGUGGGCUUUCAACCGG-GGAGCCAA--GCUGAGCUAC---AAGCUGCAGGGACCAGCAA ..(((((((....)))))))..-......((((((.((((.(((..((..((((..(((.((((..(.....)-..))))..--)))))))..)---)))).)))).).))))).. ( -42.00) >DroVir_CAF1 63188 93 - 1 AACAUUGUUCUAAAACAAUGAAAAUACAUGGUAAAA--------GAGG----AG-CCCAGGGCUCAGAGCU---GGAGCCCA--ACUGAGCGCCCGAAAGCU-----AACCAACGA ..(((((((....)))))))........((((....--------..((----.(-(.((((((((......---.)))))).--..)).)).))(....)..-----.)))).... ( -25.80) >DroSec_CAF1 47903 109 - 1 AACAUUGUUUUAAAACAAUGAA-AUACAAGCUGGUGCUGCCGCUGCUGCUGCUUCUGGUGGGCUUUCAACCGG-GGAGCCAA--GCUGAGCUAC---AAGCUGCAAGGACCAGCAA ..(((((((....)))))))..-......((((((.((((.(((..((..((((..(((.((((..(.....)-..))))..--)))))))..)---)))).)).)).)))))).. ( -41.70) >DroSim_CAF1 53553 109 - 1 AACAUUGUUUUAAAACAAUGAA-AUACAAGCUGGUGCUGCCGCUGCUGCUGCUUCUGGUGGGCUUUCAACCGG-GGAGCCAA--GCUGAGCUAC---AAGCUGCAGGGACCAGCAA ..(((((((....)))))))..-......((((((.((((.(((..((..((((..(((.((((..(.....)-..))))..--)))))))..)---)))).)))).).))))).. ( -42.00) >DroYak_CAF1 50876 93 - 1 AACAUUGUUUUAAAACAAUGAA-AUACAAGCUGCUG-----------------GCUGGUGGGCUUUCAACCGGGGGAGCCGA--GCUGAGCUAC---AAGCUGCAGGGACCAGCAA ..(((((((....)))))))..-........(((((-----------------(((....((((..(......)..))))..--.((((((...---..))).))))).)))))). ( -32.60) >DroAna_CAF1 50926 94 - 1 AACAUUGUUUUAAAACAAUGAA-AUACAAGCCGGUCAUA-----GAAGC---AA-GAGGGGGC------GCGG-AGGGGCAACGACUGAGCUAC---AUG--GCAGAGACCAACAA ..(((((((....)))))))..-.....((((((((...-----...((---..-......))------((..-....))...))))).)))..---.((--(......))).... ( -21.00) >consensus AACAUUGUUUUAAAACAAUGAA_AUACAAGCUGGUGCUG_____GAUGC___UUCUGGUGGGCUUUCAACCGG_GGAGCCAA__GCUGAGCUAC___AAGCUGCAGGGACCAGCAA ..(((((((....))))))).........((((((.........................((((((........)))))).....((((((........))).)))..)))))).. (-16.79 = -19.68 + 2.89)
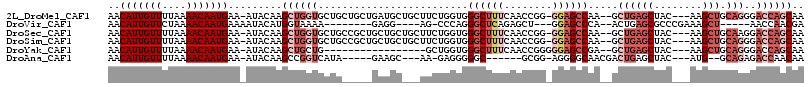
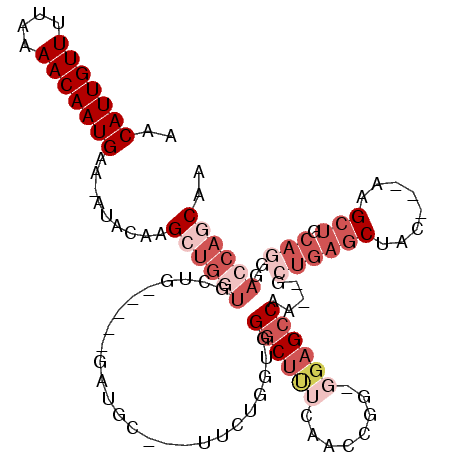
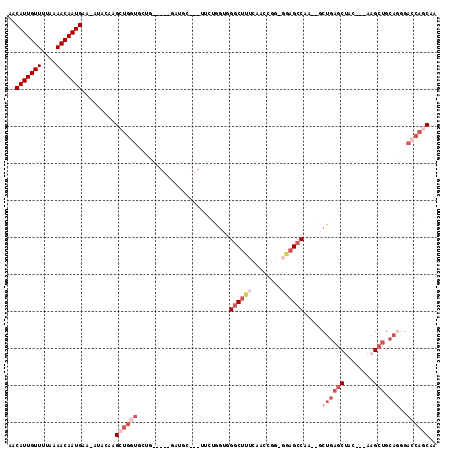
| Location | 8,619,678 – 8,619,787 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 76.52 |
| Mean single sequence MFE | -31.55 |
| Consensus MFE | -13.70 |
| Energy contribution | -14.07 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8619678 109 - 22407834 AAUCAAAGUCAGCAGCGCAGCCAAAUGGGGAAACAUUGUUUUAAAACAAUGAA-AUACAAGCUGGUGCUGCUGCUGAUGCUGCUUCUGGUGGGCUUUCAACCGG-GGAGCC .......((((((((((((.(((....(.....(((((((....)))))))..-...)....)))))).)))))))))...(((((((((.........))).)-))))). ( -37.40) >DroVir_CAF1 63217 95 - 1 AAUCAAAGCCAGUCGCGCAGCCAAAUGGGGAAACAUUGUUCUAAAACAAUGAAAAUACAUGGUAAAA--------GAGG----AG-CCCAGGGCUCAGAGCU---GGAGCC ........((((((..(.((((...((((....(((((((....)))))))....(((...)))...--------....----..-)))).))))).)).))---)).... ( -24.70) >DroSec_CAF1 47934 109 - 1 AAUCAAAGUCAGCAGUGCAGCCAAAUGGGAAAACAUUGUUUUAAAACAAUGAA-AUACAAGCUGGUGCUGCCGCUGCUGCUGCUUCUGGUGGGCUUUCAACCGG-GGAGCC .(((((((((((((((((((((.....(.....(((((((....)))))))..-...)......).)))).))))))))..)))).)))).((((..(.....)-..)))) ( -37.00) >DroSim_CAF1 53584 109 - 1 AAUCAAAGUCAGCAGCGCAGCCAAAUGGGGAAACAUUGUUUUAAAACAAUGAA-AUACAAGCUGGUGCUGCCGCUGCUGCUGCUUCUGGUGGGCUUUCAACCGG-GGAGCC .(((((((((((((((((((((.....(.....(((((((....)))))))..-...)......).)))).))))))))..)))).)))).((((..(.....)-..)))) ( -38.90) >DroYak_CAF1 50907 93 - 1 AAUCAAAGUCAGCAGCGCAGCCAAAUGGGGAAACAUUGUUUUAAAACAAUGAA-AUACAAGCUGCUG-----------------GCUGGUGGGCUUUCAACCGGGGGAGCC ...((.((((((((((....((....)).....(((((((....)))))))..-......)))))))-----------------)))..))((((..(......)..)))) ( -35.10) >DroMoj_CAF1 74699 94 - 1 AAUCAAAGUCAGUCGCGCAACCAAAUGG-GAAACAUUGUUCUAAAACAAUGAAAAUACAUGGUAAAA--------GAGG----AG-CCCAGGGCCCAGAGCC---AGAGCC ..............(.((.((((..((.-....(((((((....)))))))......))))))....--------..((----.(-((...)))))...)))---...... ( -16.20) >consensus AAUCAAAGUCAGCAGCGCAGCCAAAUGGGGAAACAUUGUUUUAAAACAAUGAA_AUACAAGCUGGUG________GAUG____UGCUGGUGGGCUUUCAACCGG_GGAGCC ............((((....((....)).....(((((((....))))))).........))))...........................((((((........)))))) (-13.70 = -14.07 + 0.36)

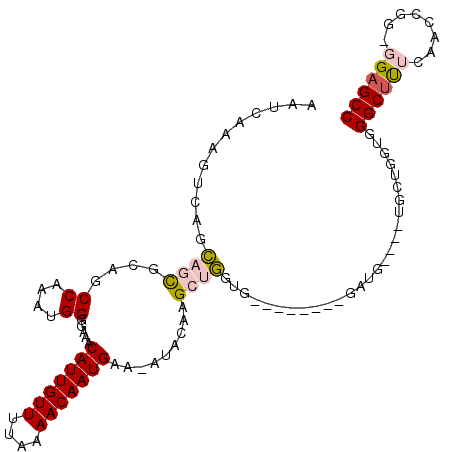
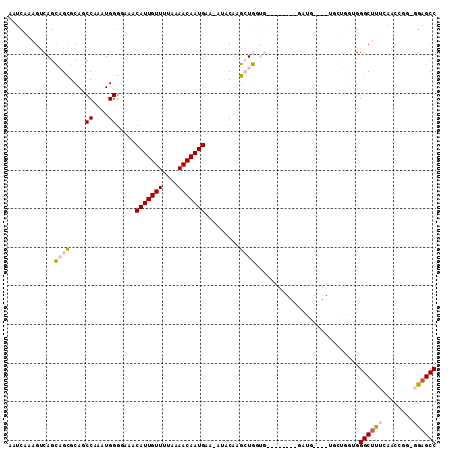
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:28 2006