
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,617,941 – 8,618,038 |
| Length | 97 |
| Max. P | 0.975423 |

| Location | 8,617,941 – 8,618,038 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.41 |
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -18.17 |
| Energy contribution | -18.47 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.975020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
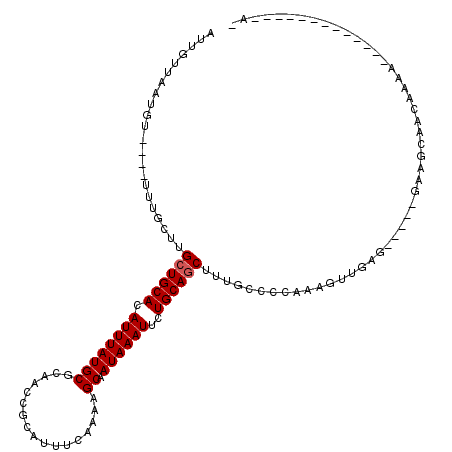
>2L_DroMel_CAF1 8617941 97 + 22407834 -U-------------UUUUGUUGCUUC-----CACAACUUUGGGGCAAAGCUGCAGAAUUUAUUGCUUUCGAAAUGCGGUUGCGCAUAAAUGUGCAGCAAGCAAA----ACAUGAACAAU -.-------------((((((((((.(-----((......))))))...((((((..(((((((((..(((.....)))..))).)))))).)))))).))))))----).......... ( -28.90) >DroVir_CAF1 61383 90 + 1 UU-------------UUUUGUUGC-----------------AGCGCAAAGCUGCAAAAUUUAUUGCUUUUGAAAUGUGGCUGCGCAUAAAUGUGCAGCAGAACAACUGCAAACAAACAGG ..-------------.((((((((-----------------(((.....)))))).......((((..(((.......((((((((....))))))))....)))..))))))))).... ( -29.60) >DroSec_CAF1 46241 97 + 1 -U-------------UUUUGUUGCUUC-----CACAACUUUGGGGCAAAGCUGCAGAAUUUAUUGCUUUAGAAAUGCGGUUGCGCAUAAAUGUGCAGCAAGCAAA----ACAUGAACAAU -.-------------((((((((((.(-----((......))))))...((((((.(.((((......)))).(((((....)))))...).)))))).))))))----).......... ( -25.30) >DroEre_CAF1 47871 97 + 1 -U-------------UUUUGUUGUUUC-----CUCAACUUUGGGGCAAAGCUGCAGAAUUUAUUGCUUUUGAAAUGCGGUUGCGCAUAAAUGUGCAGCAAGCAAA----ACAUUAACAAU -.-------------(((((((.((((-----((((....))))).)))((((((..(((((((((..(((.....)))..))).)))))).)))))).))))))----).......... ( -26.40) >DroYak_CAF1 49226 97 + 1 -U-------------UUUUGUUGUUUC-----CUCAACUUUGGAGCAAAGCUGCAGAAUUUAUUGCUUUUGAAAUGCGGUUGCGCAUAAAUGUGCAGCAAGCAAA----ACAUUAACAAU -.-------------((((((((((.(-----(........)))))...((((((..(((((((((..(((.....)))..))).)))))).)))))).))))))----).......... ( -24.90) >DroAna_CAF1 49110 110 + 1 -UUUGAUGUUUUUAUUUUUGUUGGCUUGUUGGGGAAACUUCAGCGCAAAGCUGCAGAAUUUAUUGCUUUUGAAAUGCGGUUGCGCAUAAAUGUGCACCA-----A----ACACCAACAAU -................(((((((....(((((....)....(((((..((((((...((((.......)))).)))))))))))...........)))-----)----...))))))). ( -30.30) >consensus _U_____________UUUUGUUGCUUC_____CACAACUUUGGGGCAAAGCUGCAGAAUUUAUUGCUUUUGAAAUGCGGUUGCGCAUAAAUGUGCAGCAAGCAAA____ACAUGAACAAU .................((((((((((..............)))))...((((((..(((((((((..(((.....)))..))).)))))).))))))................))))). (-18.17 = -18.47 + 0.31)


| Location | 8,617,941 – 8,618,038 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.41 |
| Mean single sequence MFE | -25.67 |
| Consensus MFE | -11.90 |
| Energy contribution | -12.06 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8617941 97 - 22407834 AUUGUUCAUGU----UUUGCUUGCUGCACAUUUAUGCGCAACCGCAUUUCGAAAGCAAUAAAUUCUGCAGCUUUGCCCCAAAGUUGUG-----GAAGCAACAAAA-------------A- ..........(----((((.(((((((......(((((....))))).......))).....(((..((((((((...))))))))..-----))))))))))))-------------.- ( -27.42) >DroVir_CAF1 61383 90 - 1 CCUGUUUGUUUGCAGUUGUUCUGCUGCACAUUUAUGCGCAGCCACAUUUCAAAAGCAAUAAAUUUUGCAGCUUUGCGCU-----------------GCAACAAAA-------------AA ....((((((.(((((.((...((((((.((((((((.................)).))))))..))))))...)))))-----------------)))))))).-------------.. ( -26.23) >DroSec_CAF1 46241 97 - 1 AUUGUUCAUGU----UUUGCUUGCUGCACAUUUAUGCGCAACCGCAUUUCUAAAGCAAUAAAUUCUGCAGCUUUGCCCCAAAGUUGUG-----GAAGCAACAAAA-------------A- ..........(----((((.(((((((......(((((....))))).......))).....(((..((((((((...))))))))..-----))))))))))))-------------.- ( -27.42) >DroEre_CAF1 47871 97 - 1 AUUGUUAAUGU----UUUGCUUGCUGCACAUUUAUGCGCAACCGCAUUUCAAAAGCAAUAAAUUCUGCAGCUUUGCCCCAAAGUUGAG-----GAAACAACAAAA-------------A- ((((((..((.----..(((((((.(((......)))))))..)))...))..))))))...((((.((((((((...)))))))).)-----))).........-------------.- ( -23.20) >DroYak_CAF1 49226 97 - 1 AUUGUUAAUGU----UUUGCUUGCUGCACAUUUAUGCGCAACCGCAUUUCAAAAGCAAUAAAUUCUGCAGCUUUGCUCCAAAGUUGAG-----GAAACAACAAAA-------------A- ((((((..((.----..(((((((.(((......)))))))..)))...))..))))))...((((.((((((((...)))))))).)-----))).........-------------.- ( -23.20) >DroAna_CAF1 49110 110 - 1 AUUGUUGGUGU----U-----UGGUGCACAUUUAUGCGCAACCGCAUUUCAAAAGCAAUAAAUUCUGCAGCUUUGCGCUGAAGUUUCCCCAACAAGCCAACAAAAAUAAAAACAUCAAA- .((((((((..----(-----((((((......(((((....))))).......))).......((.((((.....)))).)).....))))...))))))))................- ( -26.52) >consensus AUUGUUAAUGU____UUUGCUUGCUGCACAUUUAUGCGCAACCGCAUUUCAAAAGCAAUAAAUUCUGCAGCUUUGCCCCAAAGUUGAG_____GAAGCAACAAAA_____________A_ ......................((((((.((((((((.................)).))))))..))))))................................................. (-11.90 = -12.06 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:25 2006