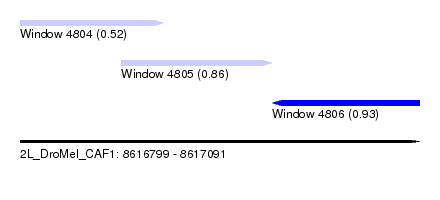
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,616,799 – 8,617,091 |
| Length | 292 |
| Max. P | 0.933123 |

| Location | 8,616,799 – 8,616,904 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 88.30 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -17.39 |
| Energy contribution | -19.83 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8616799 105 + 22407834 UUUAUUAACAGAUACAACUUUCUUUAUAUGGGGUUUAAUCCUAAUGCUACCAUAGAUACAAACCAAAAGAUUUAUGGAGCAAUCUCGCUGUCAGUUUCUCCUGU---------A .......((((....((((..(......((((((...)))))).((((.((((((((............))))))))))))........)..))))....))))---------. ( -19.80) >DroSec_CAF1 45088 114 + 1 UUUAUUAUCAGAUAGAUACUUCUUUAUAUGAGGUUUAAUCCUAAUGCUGCCAUAGAUUUAAACCAAAAGAUUUAUGGAGCAAUCACGCUGUCAGUUUCUCCUGUAAAGUGUGUA ..........(((.((.(((((.......))))))).)))....((((.((((((((((........))))))))))))))..((((((..(((......)))...)))))).. ( -28.60) >DroSim_CAF1 50774 114 + 1 UUUAUUAUCAGAUAGAUACUUCUUUAUAUGAGCUUUAAUCCUAAUGCUGCCAUAGAUUUAAACCAAAAGAUUUAUGGAGCAAUCACGCUGUCAGUUUCUCCUGUAAAGUGUGUA .......(((.(((((......))))).))).............((((.((((((((((........))))))))))))))..((((((..(((......)))...)))))).. ( -26.60) >consensus UUUAUUAUCAGAUAGAUACUUCUUUAUAUGAGGUUUAAUCCUAAUGCUGCCAUAGAUUUAAACCAAAAGAUUUAUGGAGCAAUCACGCUGUCAGUUUCUCCUGUAAAGUGUGUA (((((....(((........)))....)))))............((((.((((((((((........))))))))))))))..((((((..(((......)))...)))))).. (-17.39 = -19.83 + 2.45)

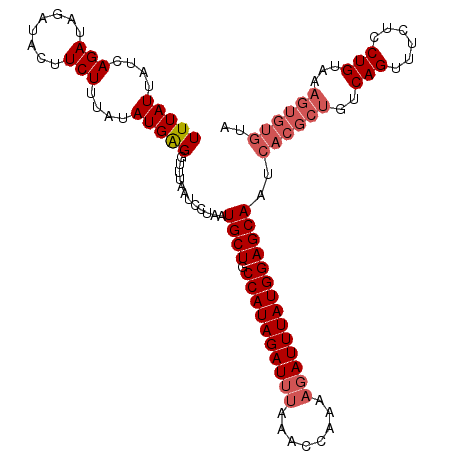

| Location | 8,616,873 – 8,616,983 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.58 |
| Mean single sequence MFE | -28.60 |
| Consensus MFE | -14.66 |
| Energy contribution | -16.10 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8616873 110 + 22407834 UGGAGCAAUCUCGCUGUCAGUUUCUCCUGU---------ACUAGUUAGAUAGUUGGAUUUACCCCAUCUCCCUUUUGAUAAUGUUCCUCUAAAAU-UUCCCGAGAUGGUCUUCUAUCUCG .((((((((((.((((((((......))).---------)).))).))))((.(((.......))).))............))))))........-....((((((((....)))))))) ( -27.60) >DroSec_CAF1 45162 118 + 1 UGGAGCAAUCACGCUGUCAGUUUCUCCUGUAAAGUGUGUACUA-UUGGUUAGUUGGAUUCC-CCCACCUGCCUUUUGGUGAUUUUCCUUUUAAAUUUUCCCGAGAUGGUCUUCUAUCUCG .((((.(((((((((..(((......)))...)))))..((((-..(((.((.(((.....-.))).)))))...)))))))))))).............((((((((....)))))))) ( -33.10) >DroSim_CAF1 50848 118 + 1 UGGAGCAAUCACGCUGUCAGUUUCUCCUGUAAAGUGUGUACUA-UUGGAUAGUUGGAUUCCCCCCACCUGCCUUUUGGUGAUGUUCCUCUGAAAU-UUCCCGAGAUGGUCUUCUAUCUCG .((((((..((((((..(((......)))...))))))(((((-..((.(((.(((.......))).)))))...))))).))))))........-....((((((((....)))))))) ( -37.60) >DroEre_CAF1 46808 102 + 1 UGGC---------------GUUUCUCCCGCAAAGUUUAUACCA-UCGGAUAGUUGGACUUACCCCACAUCCC-UCUGGUAAUAUUCCCCCCAAAU-UUCCCGAGAUGGUAUUCUAUCUCG ..((---------------(.......)))........(((((-..((((.((.((......)).)))))).-..)))))...............-....((((((((....)))))))) ( -24.40) >DroYak_CAF1 48077 117 + 1 UGGAGUAAGCCCGCAGACAGUUUCGCCUGUAAAGUGUUUACUA-UCGAAUAGCUAGAUUUACCCUACAUCCC-UUUGGUAAUAUUUCUCUUAAAU-UUCCCGAGAUGGUAUUCUAUCGCG .(((((((((.(....((((......))))...).))))))).-)).....((((((..((((...((....-..))........((((......-.....)))).)))).))))..)). ( -20.30) >consensus UGGAGCAAUCACGCUGUCAGUUUCUCCUGUAAAGUGUGUACUA_UUGGAUAGUUGGAUUUACCCCACCUCCCUUUUGGUAAUAUUCCUCUUAAAU_UUCCCGAGAUGGUCUUCUAUCUCG .((((((..........(((......))).........(((((...((.....(((.......)))....))...))))).)))))).............((((((((....)))))))) (-14.66 = -16.10 + 1.44)



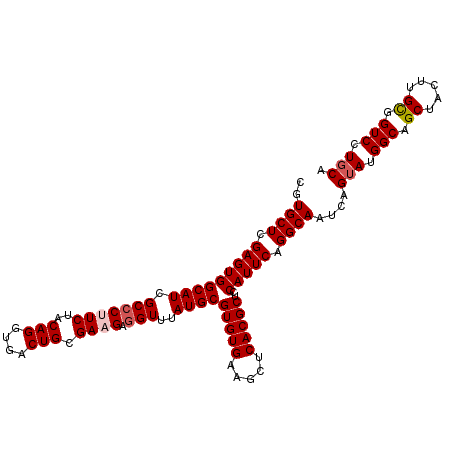
| Location | 8,616,983 – 8,617,091 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -39.88 |
| Consensus MFE | -37.06 |
| Energy contribution | -37.70 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8616983 108 - 22407834 CGUGCUCGAGUGGCAUCGCCCUUCUACAGGUGACUGCGAAGAGGUUUAUGCGUGUGAAUCUCACGCUCCAUUCAGGCAAUCAGUAUGGCAGCUACUUGUGGUCCUGCA ..(((..(((((((((.(((((((..(((....))).)))).)))..))))(((((.....)))))..)))))..)))....(((.(((.((.....)).))).))). ( -39.70) >DroSec_CAF1 45280 108 - 1 CGUGCUCGAGUGGCAUCGCCCUUCUACAGGUGACUGCGAAGAGGUUUAUGCGUGUGAAGCUCACGCUCCAUUCAGGCAAUCAGUAUGGCAGCUACUUGCGGUCCUGCA ..(((..(((((((((.(((((((..(((....))).)))).)))..))))(((((.....)))))..)))))..)))....(((.(((.((.....)).))).))). ( -41.30) >DroSim_CAF1 50966 108 - 1 CGUGCUCGAGUGGCAUCGCCCUUCUACAGGUGACUGCGAAGAGGUUUAUGCGUGUGAAGCUCACGCUCCAUUCAGGCAAUCAGUAUGGCAGCUACUUGCGGUCCUGCA ..(((..(((((((((.(((((((..(((....))).)))).)))..))))(((((.....)))))..)))))..)))....(((.(((.((.....)).))).))). ( -41.30) >DroEre_CAF1 46910 107 - 1 CGUGCUCGAGUGGCAUCGC-CGUCUACAGGUGACUGCGAAGAGGUUUAUGCGUGUGAAGCUCACGCUACAUUCGGGCAAUCGGUAUGGCAGCUACUUGCGGUCCUGCA ..((((((((((((((.((-(.(((.(((....)))...))))))..))))(((((.....)))))..))))))))))....(((.(((.((.....)).))).))). ( -43.20) >DroYak_CAF1 48194 108 - 1 CGUGCUCGAGUGGCAUCGCCCGUCUACAGGUGACUGCGAAGAGGUUUAUGCGUGUGAAGCUCACGCUACAUUCAGGCAAUCAGUAUGGCAGCUACUUGCGGUCCUUCA ..(((..(((((((((.((((.((..(((....))).)).).)))..))))(((((.....)))))..)))))..)))........(((.((.....)).)))..... ( -33.90) >consensus CGUGCUCGAGUGGCAUCGCCCUUCUACAGGUGACUGCGAAGAGGUUUAUGCGUGUGAAGCUCACGCUCCAUUCAGGCAAUCAGUAUGGCAGCUACUUGCGGUCCUGCA ..((((.(((((((((.(((((((..(((....))).)))).)))..))))(((((.....)))))..))))).))))....(((.(((.((.....)).))).))). (-37.06 = -37.70 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:21 2006