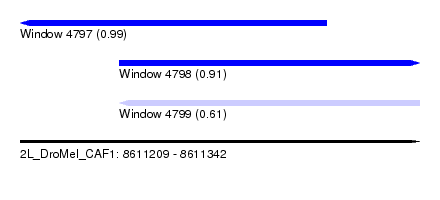
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,611,209 – 8,611,342 |
| Length | 133 |
| Max. P | 0.986530 |

| Location | 8,611,209 – 8,611,311 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.70 |
| Mean single sequence MFE | -49.93 |
| Consensus MFE | -23.56 |
| Energy contribution | -24.62 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.31 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.47 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
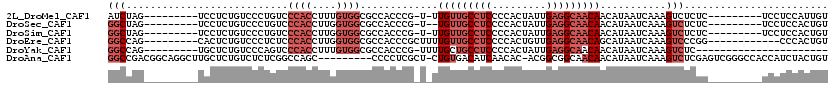
>2L_DroMel_CAF1 8611209 102 - 22407834 UUGCCUCAAUAGUGGGGAGGCAACAA-A-CGGGUGGCGCCACAAAGGUGGGACAGGGACAGAGGA---------CUAGAUCUCGGCGGUCCGUCUCUGUUCGUCGC-------CGGUUUA (((((((.........))))))).((-(-(.(((((((((((....))))(((((((((...(((---------((.(....)...))))))))))))))))))))-------).)))). ( -46.40) >DroSec_CAF1 39510 101 - 1 UUGCCUCAAUAGUGGGGAGGCAACA--A-CGGGUGGCGCCACCAAGGUGGGACAGGGACAGAGGA---------CUAGCCCUCGGCGGUCCGUCUCUGUUCGUCGC-------CGGUUUA (((((((.........))))))).(--(-(.(((((((((((....))))(((((((((...(((---------((.(((...)))))))))))))))))))))))-------).))).. ( -50.80) >DroSim_CAF1 45231 102 - 1 UUGCCUCAAUAGUGGGGAGGCAACAA-A-CGGGUGGCGCCACCAAGGUGGGACAGGGACAGAGGA---------CUAGCCCUCGGCGGUCCGUCUCUGUUCGUCGC-------CGGUUUA (((((((.........))))))).((-(-(.(((((((((((....))))(((((((((...(((---------((.(((...)))))))))))))))))))))))-------).)))). ( -52.30) >DroEre_CAF1 41256 104 - 1 UUGCCUCAACAGUGGGGAGGCAACAAAAGCGGGUGGCGCCACCAAGGUGGGAGAGGGACAGAGUG---------CUGGCCCACGGCGCUCCAUCUCUGUUCGUCGC-------CGGUUUA (((((((..(...)..)))))))...((((.(((((((((((....))))((.(((((..(((((---------(((.....))))))))..))))).))))))))-------).)))). ( -52.10) >DroYak_CAF1 42077 103 - 1 UUGCCUCAAUAGUGGGGAGGCAGCAAAA-CGGGUGGCGCCACAAAGGUGGGACUGGGACAGAGCA---------CUGGCCCACGGCGCUCUGUCUCAGUUCGUCGU-------CGGUUUA (((((((.........)))))))..(((-(.(..((((((((....))))((((((((((((((.---------(((.....))).))))))))))))))))))..-------).)))). ( -53.10) >DroAna_CAF1 40196 109 - 1 UUGCCGCCGU-GUGUUGAUGUCACAG-AGCGAGGGG---------GCUGGCCGAGAGACAGAGCAAGCCUGCCGUCGGCCGUUGGCGGUGCGUCUCUGUUCGUCCUCGGCACUCGGUUUC ....(((..(-(((.......)))).-.)))..(..---------((((((((((.(((((((...((((((((........)))))).))..)))))))....))))))...))))..) ( -44.90) >consensus UUGCCUCAAUAGUGGGGAGGCAACAA_A_CGGGUGGCGCCACCAAGGUGGGACAGGGACAGAGGA_________CUAGCCCUCGGCGGUCCGUCUCUGUUCGUCGC_______CGGUUUA (((((((.........))))))).........((((((((((....))))(((((((((...(((............(((...)))..)))))))))))))))))).............. (-23.56 = -24.62 + 1.06)

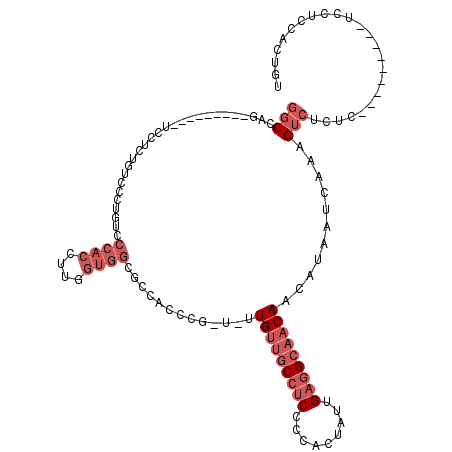
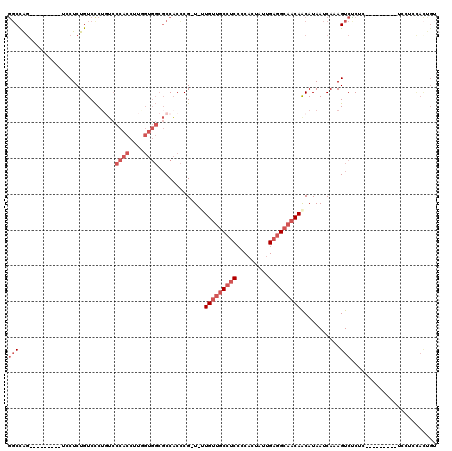
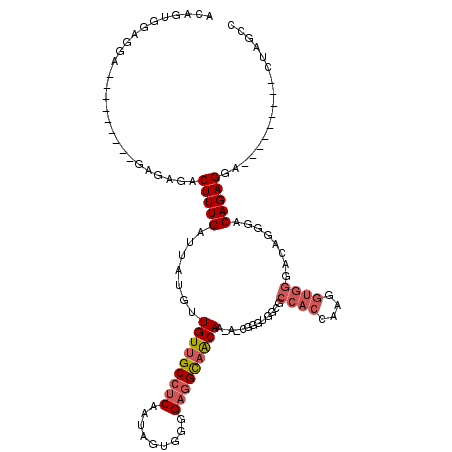
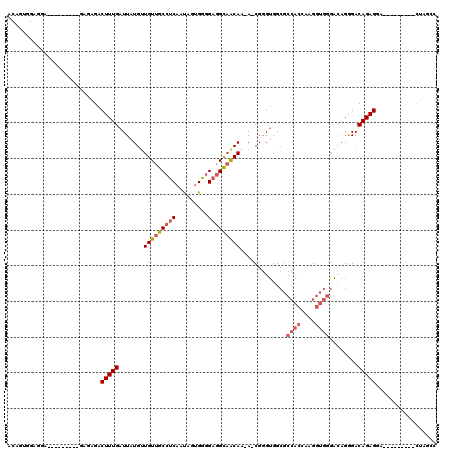
| Location | 8,611,242 – 8,611,342 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.58 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -12.47 |
| Energy contribution | -14.30 |
| Covariance contribution | 1.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8611242 100 + 22407834 AUCUAG---------UCCUCUGUCCCUGUCCCACCUUUGUGGCGCCACCCG-U-UUGUUGCCUCCCCACUAUUGAGGCAACAACAUAAUCAAAGUCUCUC---------UCCUCCAUUGU ......---------............((.((((....)))).)).....(-(-(.((((((((.........)))))))))))................---------........... ( -19.20) >DroSec_CAF1 39543 99 + 1 GGCUAG---------UCCUCUGUCCCUGUCCCACCUUGGUGGCGCCACCCG-U--UGUUGCCUCCCCACUAUUGAGGCAACAACAUAAUCAAAGUCUCUC---------UCCUCCACUGU ((((..---------............((.((((....)))).)).....(-(--(((((((((.........)))))))))))........))))....---------........... ( -28.30) >DroSim_CAF1 45264 100 + 1 GGCUAG---------UCCUCUGUCCCUGUCCCACCUUGGUGGCGCCACCCG-U-UUGUUGCCUCCCCACUAUUGAGGCAACAACAUAAUCAAAGUCUCUC---------UCCUCCACUGU ((((..---------............((.((((....)))).)).....(-(-(.((((((((.........)))))))))))........))))....---------........... ( -24.50) >DroEre_CAF1 41289 99 + 1 GGCCAG---------CACUCUGUCCCUCUCCCACCUUGGUGGCGCCACCCGCUUUUGUUGCCUCCCCACUGUUGAGGCAACAGCAUAAUCAAAGUCCCGG------------CCCACUGU ((((.(---------.(((.((...............((((....)))).(((...((((((((.........))))))))))).....)).))).).))------------))...... ( -30.40) >DroYak_CAF1 42110 88 + 1 GGCCAG---------UGCUCUGUCCCAGUCCCACCUUUGUGGCGCCACCCG-UUUUGCUGCCUCCCCACUAUUGAGGCAACAACAUAAUCAAAGUCUC---------------------- ((((((---------....)))........((((....)))).))).....-..(((.((((((.........)))))).)))...............---------------------- ( -19.20) >DroAna_CAF1 40236 109 + 1 GGCCGACGGCAGGCUUGCUCUGUCUCUCGGCCAGC---------CCCCUCGCU-CUGUGACAUCAACAC-ACGGCGGCAACAACAUAAUCAAAGUCUCGAGUCGGGCCACCAUCUACUGU ((((((.((((((....).)))))..)))))).((---------((.((((((-.((((.......)))-).)))(....).................)))..))))............. ( -38.00) >consensus GGCCAG_________UCCUCUGUCCCUGUCCCACCUUGGUGGCGCCACCCG_U_UUGUUGCCUCCCCACUAUUGAGGCAACAACAUAAUCAAAGUCUCUC_________UCCUCCACUGU (((...........................((((....)))).............(((((((((.........)))))))))...........)))........................ (-12.47 = -14.30 + 1.83)
| Location | 8,611,242 – 8,611,342 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.58 |
| Mean single sequence MFE | -32.65 |
| Consensus MFE | -19.21 |
| Energy contribution | -20.10 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8611242 100 - 22407834 ACAAUGGAGGA---------GAGAGACUUUGAUUAUGUUGUUGCCUCAAUAGUGGGGAGGCAACAA-A-CGGGUGGCGCCACAAAGGUGGGACAGGGACAGAGGA---------CUAGAU ...........---------......(((((.(..((((((((((((.........))))))))..-.-.........((((....))))))))..).)))))..---------...... ( -25.90) >DroSec_CAF1 39543 99 - 1 ACAGUGGAGGA---------GAGAGACUUUGAUUAUGUUGUUGCCUCAAUAGUGGGGAGGCAACA--A-CGGGUGGCGCCACCAAGGUGGGACAGGGACAGAGGA---------CUAGCC ........((.---------.((...(((((....((((((((((((.........)))))))))--)-))((((....))))...............)))))..---------))..)) ( -33.00) >DroSim_CAF1 45264 100 - 1 ACAGUGGAGGA---------GAGAGACUUUGAUUAUGUUGUUGCCUCAAUAGUGGGGAGGCAACAA-A-CGGGUGGCGCCACCAAGGUGGGACAGGGACAGAGGA---------CUAGCC ........((.---------.((...(((((.(..((((((((((((.........))))))))..-.-..((((....)))).......))))..).)))))..---------))..)) ( -30.50) >DroEre_CAF1 41289 99 - 1 ACAGUGGG------------CCGGGACUUUGAUUAUGCUGUUGCCUCAACAGUGGGGAGGCAACAAAAGCGGGUGGCGCCACCAAGGUGGGAGAGGGACAGAGUG---------CUGGCC ......((------------((((.((((((....((((((((((((..(...)..))))))))...))))((((....))))...............)))))).---------)))))) ( -42.50) >DroYak_CAF1 42110 88 - 1 ----------------------GAGACUUUGAUUAUGUUGUUGCCUCAAUAGUGGGGAGGCAGCAAAA-CGGGUGGCGCCACAAAGGUGGGACUGGGACAGAGCA---------CUGGCC ----------------------..(.(((((......((((((((((.........))))))))))..-(.(((..((((.....))))..))).)..)))))).---------...... ( -28.60) >DroAna_CAF1 40236 109 - 1 ACAGUAGAUGGUGGCCCGACUCGAGACUUUGAUUAUGUUGUUGCCGCCGU-GUGUUGAUGUCACAG-AGCGAGGGG---------GCUGGCCGAGAGACAGAGCAAGCCUGCCGUCGGCC .(((((.((((((((.((((..((........))..))))..))))))))-.))))).........-(((......---------)))((((((..(.(((.(....)))))..)))))) ( -35.40) >consensus ACAGUGGAGGA_________GAGAGACUUUGAUUAUGUUGUUGCCUCAAUAGUGGGGAGGCAACAA_A_CGGGUGGCGCCACCAAGGUGGGACAGGGACAGAGGA_________CUAGCC ..........................(((((.......(((((((((.........))))))))).............((((....))))........)))))................. (-19.21 = -20.10 + 0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:14 2006