
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,606,250 – 8,606,360 |
| Length | 110 |
| Max. P | 0.927065 |

| Location | 8,606,250 – 8,606,360 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 84.30 |
| Mean single sequence MFE | -29.08 |
| Consensus MFE | -24.26 |
| Energy contribution | -25.30 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
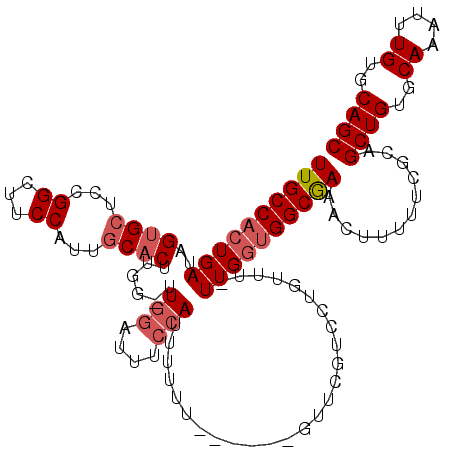
>2L_DroMel_CAF1 8606250 110 + 22407834 UUGCCACUGAUAGUGCUCCGGCUUCCAUUGCACUUGG-UUGGAUUUCCAUUUUUU------GUUCGUAGUGUUUUUUGGUGGCGAAACUUAUUCGCAGCUGUGCAAAUUUGUGCAGC (((((((..(.((..((.(((...(((.......)))-.(((....)))......------..))).))..))..)..)))))))............((((..((....))..)))) ( -32.40) >DroSec_CAF1 34605 108 + 1 UUGCCACUGAUAGUGCUCCGGCUUCCAUUGCACUUAG-UUGGAUUUCCAUUUUUU-------UUCGUCCUGUUU-UUGGUGGCGAAACUUUUUCGCAGCUGUGCAAAUUUGUGCAGC (((((((..(.(((((...((...))...)))))...-.(((....)))......-------............-)..)))))))............((((..((....))..)))) ( -30.80) >DroSim_CAF1 40296 109 + 1 UUGCCACUGAUAGUGCUCCGGCUUCCAUUGCACUUGG-UUGGAUUUCCAUUUUUU------GUUCGUCCUGUUU-UUGGUGGCGAAACUUUUUCGCAGCUGUGCAAAUUUGUGCAGC (((((((..(.(((((...((...))...)))))...-..((((...((.....)------)...)))).....-)..)))))))............((((..((....))..)))) ( -31.80) >DroYak_CAF1 36988 115 + 1 UUGCCAGUGAUAGUGCUCGGGCUUCCAUUGCACUUGC-UUGGAUUUCCAUUUUUUAUUUUUUUUCAUCCUGUUU-UUGGUGGCGAAACUUUUUUGCAGCUGUGCAAAUUUGUGCAGC ..(((((.(((((.....(((..((((..((....))-.))))..)))((.....))...........))))).-))))).(((((.....))))).((((..((....))..)))) ( -29.20) >DroAna_CAF1 35784 115 + 1 UUGCCCCUGAUAGCGCACGGGUUUCCAUUCCUCCUAUAUUUUUUUUCUAUUUUCCAUUUUUUUUGUUUUUACGU-UUGGUGGCAAAACUU-UUCGCAGCUGUGCAAAUUUGUGCAGC (((((((....((((...(((.......))).......................((.......))......)))-).)).))))).....-......((((..((....))..)))) ( -21.20) >consensus UUGCCACUGAUAGUGCUCCGGCUUCCAUUGCACUUGG_UUGGAUUUCCAUUUUUU______GUUCGUCCUGUUU_UUGGUGGCGAAACUUUUUCGCAGCUGUGCAAAUUUGUGCAGC ((((((((((..((((...((...))...))))......(((....)))..........................))))))))))............((((..((....))..)))) (-24.26 = -25.30 + 1.04)

| Location | 8,606,250 – 8,606,360 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 84.30 |
| Mean single sequence MFE | -23.62 |
| Consensus MFE | -17.26 |
| Energy contribution | -18.70 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
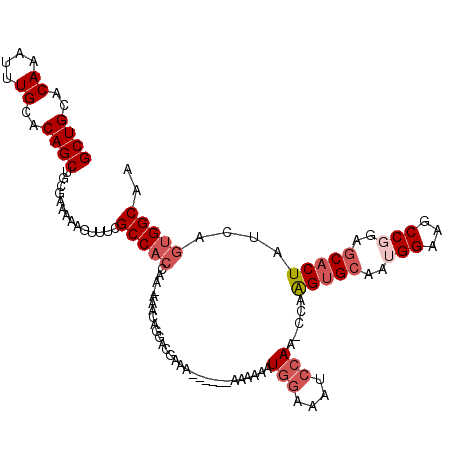
>2L_DroMel_CAF1 8606250 110 - 22407834 GCUGCACAAAUUUGCACAGCUGCGAAUAAGUUUCGCCACCAAAAAACACUACGAAC------AAAAAAUGGAAAUCCAA-CCAAGUGCAAUGGAAGCCGGAGCACUAUCAGUGGCAA ((((..((....))..))))...(((.....)))(((((.................------......(((....))).-...(((((..(((...)))..)))))....))))).. ( -25.80) >DroSec_CAF1 34605 108 - 1 GCUGCACAAAUUUGCACAGCUGCGAAAAAGUUUCGCCACCAA-AAACAGGACGAA-------AAAAAAUGGAAAUCCAA-CUAAGUGCAAUGGAAGCCGGAGCACUAUCAGUGGCAA ((..(.....((((((....))))))....(((((((.....-.....)).))))-------).....(((....))).-...(((((..(((...)))..)))))....)..)).. ( -26.30) >DroSim_CAF1 40296 109 - 1 GCUGCACAAAUUUGCACAGCUGCGAAAAAGUUUCGCCACCAA-AAACAGGACGAAC------AAAAAAUGGAAAUCCAA-CCAAGUGCAAUGGAAGCCGGAGCACUAUCAGUGGCAA ((..(.....((((((....))))))...(((.((((.....-.....)).)))))------......(((....))).-...(((((..(((...)))..)))))....)..)).. ( -25.20) >DroYak_CAF1 36988 115 - 1 GCUGCACAAAUUUGCACAGCUGCAAAAAAGUUUCGCCACCAA-AAACAGGAUGAAAAAAAAUAAAAAAUGGAAAUCCAA-GCAAGUGCAAUGGAAGCCCGAGCACUAUCACUGGCAA ((((((......))))(((.(((.......(((((((.....-.....)).)))))............(((....))).-)))(((((..(((....))).)))))....))))).. ( -23.00) >DroAna_CAF1 35784 115 - 1 GCUGCACAAAUUUGCACAGCUGCGAA-AAGUUUUGCCACCAA-ACGUAAAAACAAAAAAAAUGGAAAAUAGAAAAAAAAUAUAGGAGGAAUGGAAACCCGUGCGCUAUCAGGGGCAA ((((..((....))..))))......-.....(((((.((..-..((....))..............................((.((.((((....))))...)).)).))))))) ( -17.80) >consensus GCUGCACAAAUUUGCACAGCUGCGAAAAAGUUUCGCCACCAA_AAACAGGACGAAA______AAAAAAUGGAAAUCCAA_CCAAGUGCAAUGGAAGCCGGAGCACUAUCAGUGGCAA ((((..((....))..))))..............(((((.............................(((....))).....(((((..(((...)))..)))))....))))).. (-17.26 = -18.70 + 1.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:09 2006