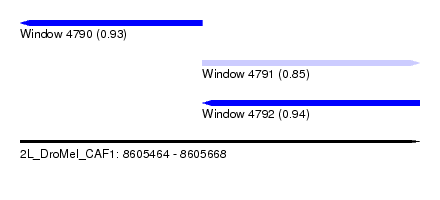
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,605,464 – 8,605,668 |
| Length | 204 |
| Max. P | 0.941440 |

| Location | 8,605,464 – 8,605,557 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.07 |
| Mean single sequence MFE | -41.42 |
| Consensus MFE | -26.49 |
| Energy contribution | -26.64 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8605464 93 - 22407834 CAUUCAGGGGUUGGC------------CAUCGGUUGGCUUUUGGGUA--------------CUGGCC-CAGCAGGCUAGCCAACUUGAUGUAUUGCCGAGGGCGUAACCCCAACUGGACU ......(((((((((------------(...(((((((((((((((.--------------...)))-))).)))))))))..((((.........))))))).)))))))......... ( -41.60) >DroSec_CAF1 33866 94 - 1 CAUUCAGGGGUUGGC------------CAUCGGUUGGCUUUUGGGUA--------------CUGGCCACAGCAGCGCAGCCAACUUGAUGGAUUGCCAAGGGCGUAACCCCAACUGGACU ......(((((((.(------------((((((((((((...(.((.--------------(((....)))..)).)))))))).))))))...(((...))).)))))))......... ( -37.00) >DroSim_CAF1 39560 94 - 1 CAUUCAGGGGUUGGC------------CAUCGGUUGGCUUUUGGGUA--------------CUGGCCACGGCAGCGCAGGCAACUUGAUGGAUUGCCGAGGGCGUAACCCCAACUGGACU ......(((((((((------------(.(((((...((.(..(((.--------------(((((.......)).)))...)))..).))...))))).))).)))))))......... ( -37.00) >DroEre_CAF1 35569 99 - 1 CAUUCAGGGGUUGGC------------CAUCGGUUGGCUUCUGGGUA-------CUGGGUACUGGCCACA-CAGC-AGGCCAACUUGAUGGAUUGCCGAGGGCGUAACCCCGACUGGACU ((.((.(((((((.(------------((((((((((((((((.((.-------...(((....))))).-))).-)))))))).))))))...(((...))).))))))))).)).... ( -41.10) >DroYak_CAF1 36177 107 - 1 CAUUCAGGGGUUGGC------------CAUCGGUUGGCUUUUGGGUAUUGGGUCCUGGGUACUGGCCACAGCAGC-AGGCCAACUUGAUGGAUUGCCGAGGGCGUAACCCCAACUGGACU ......(((((((((------------(.(((((...((.(..(((....((.((((....(((....)))...)-))))).)))..).))...))))).))).)))))))......... ( -42.90) >DroAna_CAF1 34976 107 - 1 CAUUCGGGGGGUGGUCAUCGAGGUGGUCAUCGGCUGGCUCU-GGCU--------CUGGGUCCAGACCAUU-C---UGAGCCAGCUUGAUGGAUUGCCAGGGGCGCAACCCAAACUAGAUU .....(((..(((.((.....((..((((((((((((((((-((.(--------(((....)))))))..-.---.)))))))).)))).)))..))..)).)))..))).......... ( -48.90) >consensus CAUUCAGGGGUUGGC____________CAUCGGUUGGCUUUUGGGUA______________CUGGCCACAGCAGC_AAGCCAACUUGAUGGAUUGCCGAGGGCGUAACCCCAACUGGACU ......(((((((..............((((((((((((.((((((..................))).)))......))))))).)))))....(((...))).)))))))......... (-26.49 = -26.64 + 0.14)
| Location | 8,605,557 – 8,605,668 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 93.90 |
| Mean single sequence MFE | -24.28 |
| Consensus MFE | -22.38 |
| Energy contribution | -22.14 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846310 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8605557 111 + 22407834 CAUUAGCCAUAACUCUCAUUAUGUGGCAUCCGAUCCGGACCGGGGUCUUAGCUCCGCAGCCCAGCGACCCAUUCAAUCACGCUAUAGCGCUCCC-AAAUUUCCCAACGCUCC .....((((((..........)))))).........(((.(((((((...(((....))).....))))).........(((....))).....-...........)).))) ( -23.80) >DroSec_CAF1 33960 111 + 1 CAUUAGCCAUAACUCUCAUUAUGUGGCAUCCGAUCCGGACCCGGGUCAUAGCUCCAGAGCCCAGCGACCCAUUCAAUCACGCUAUAGCGCCCCC-AAAUUUCCCACCGCUCC .....((((((..........))))))....((((((....)))))).........((((.....((.....)).....(((....))).....-............)))). ( -24.40) >DroSim_CAF1 39654 111 + 1 CAUUAGCCAUAACUCUCAUUAUGUGGCAUCCGAUCCGGACCCGGGUCAUAGCUCCAGAGCCCAGCGACCCAUUCAAUCACGCUAUAGCGCCCUC-AAAUUUCCCACCGCUCC .....((((((..........))))))....((((((....)))))).........((((.....((.....)).....(((....))).....-............)))). ( -24.40) >DroEre_CAF1 35668 112 + 1 CAUUAGCCAUAACUCUCAUUAUGUGGCAUCCGAUCCGGACCCGGGUCAUAGCUCCUGAGUCCAGCGACCCAUUCAAUCACGCCGUAGCGCCCCCGAAAAUUUCCACCGCUCC .....((((((..........)))))).........((((.((((........)))).))))((((.............(((....))).....((.....))...)))).. ( -24.10) >DroYak_CAF1 36284 111 + 1 CAUUAGCCAUAACUCUCAUUAUGUGGCAUCCGAUCCGGACCCGGGUCAUAGCUCCAAAGUCUAGCGACCCAUUCAAUCACGCUAUAGCGCCCCC-AAAAUUUCCACCGCUCC .....((((((..........))))))....((..(((....(((((.(((((....)).)))..))))).........(((....))).....-..........))).)). ( -24.70) >consensus CAUUAGCCAUAACUCUCAUUAUGUGGCAUCCGAUCCGGACCCGGGUCAUAGCUCCAGAGCCCAGCGACCCAUUCAAUCACGCUAUAGCGCCCCC_AAAUUUCCCACCGCUCC .....((((((..........)))))).((((...))))...(((((...(((....))).....))))).........(((....)))....................... (-22.38 = -22.14 + -0.24)


| Location | 8,605,557 – 8,605,668 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 93.90 |
| Mean single sequence MFE | -39.54 |
| Consensus MFE | -32.76 |
| Energy contribution | -32.80 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8605557 111 - 22407834 GGAGCGUUGGGAAAUUU-GGGAGCGCUAUAGCGUGAUUGAAUGGGUCGCUGGGCUGCGGAGCUAAGACCCCGGUCCGGAUCGGAUGCCACAUAAUGAGAGUUAUGGCUAAUG ..(((..(((...(((.-.(..((((....))))..)..)))(((((....((((....))))..)))))..(((((...))))).)))((((((....))))))))).... ( -38.50) >DroSec_CAF1 33960 111 - 1 GGAGCGGUGGGAAAUUU-GGGGGCGCUAUAGCGUGAUUGAAUGGGUCGCUGGGCUCUGGAGCUAUGACCCGGGUCCGGAUCGGAUGCCACAUAAUGAGAGUUAUGGCUAAUG ..((((((.....(((.-.(..((((....))))..)..)))((((((...((((....)))).))))))..(((((...)))))))).((((((....))))))))).... ( -40.50) >DroSim_CAF1 39654 111 - 1 GGAGCGGUGGGAAAUUU-GAGGGCGCUAUAGCGUGAUUGAAUGGGUCGCUGGGCUCUGGAGCUAUGACCCGGGUCCGGAUCGGAUGCCACAUAAUGAGAGUUAUGGCUAAUG ..((((((.....(((.-.(..((((....))))..)..)))((((((...((((....)))).))))))..(((((...)))))))).((((((....))))))))).... ( -40.50) >DroEre_CAF1 35668 112 - 1 GGAGCGGUGGAAAUUUUCGGGGGCGCUACGGCGUGAUUGAAUGGGUCGCUGGACUCAGGAGCUAUGACCCGGGUCCGGAUCGGAUGCCACAUAAUGAGAGUUAUGGCUAAUG ..((((((.......(((((..((((....))))..)))))(((((((.(((.((....)))))))))))).(((((...)))))))).((((((....))))))))).... ( -41.30) >DroYak_CAF1 36284 111 - 1 GGAGCGGUGGAAAUUUU-GGGGGCGCUAUAGCGUGAUUGAAUGGGUCGCUAGACUUUGGAGCUAUGACCCGGGUCCGGAUCGGAUGCCACAUAAUGAGAGUUAUGGCUAAUG ..((((((......((.-.(..((((....))))..)..))(((((((.(((.((....)))))))))))).(((((...)))))))).((((((....))))))))).... ( -36.90) >consensus GGAGCGGUGGGAAAUUU_GGGGGCGCUAUAGCGUGAUUGAAUGGGUCGCUGGGCUCUGGAGCUAUGACCCGGGUCCGGAUCGGAUGCCACAUAAUGAGAGUUAUGGCUAAUG ..(((..(((............((((....))))........((((((.(((.((....)))))))))))..(((((...))))).)))((((((....))))))))).... (-32.76 = -32.80 + 0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:07 2006