| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
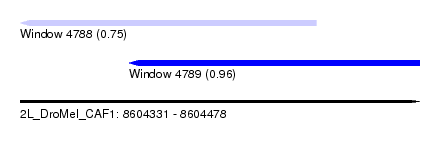
| Location | 8,604,331 – 8,604,478 |
| Length | 147 |
| Max. P | 0.959188 |

| Location | 8,604,331 – 8,604,440 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.74 |
| Mean single sequence MFE | -22.12 |
| Consensus MFE | -12.90 |
| Energy contribution | -13.10 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8604331 109 - 22407834 CUCC----CCCUUUUGAGUGUGUUGACCAACAAUGUUGGUCACAACAUGUACA--CACAACCCGCCCCA-----UACCCCGUCGUCCUGGCCCUUCUACUCGUCCUUCCACUAACAAGAA ....----...(((((.((((((((((((((...))))))))........)))--))).....(((...-----.((......))...))).......................))))). ( -20.90) >DroSec_CAF1 32753 96 - 1 CUCC----CCCUUUUGAGUCUGUUGACCAACAAUGUUGGUCACAACAUGUACA--CACAG----C-CCA-----UGUCCUG--------GCCCUUCUACUCGUCCUUCCACUAACAAGAA ....----.......((((.(((.(((((((...))))))))))(((((..(.--....)----.-.))-----)))....--------........))))................... ( -17.30) >DroSim_CAF1 38438 102 - 1 CUCC----CCCUUUUGAGUGUGUUGACCAACAAUGUUGGUCACAACAUGUACA--CACAGCUCGC-CCA-----UACCCCGUC------GUCCUUCUACUCGUCCUUCCACUAACAAGAA ....----...(((((.((((((((((((((...))))))))........)))--))).......-...-----......(.(------(..........)).)..........))))). ( -18.00) >DroEre_CAF1 34498 104 - 1 CUCC----CACUUUUGAGUGUGUUGACCAACAAUGUUGGUCACAACAUGUACACACACAG----C-CCA-------GCCCCUCGUCCUGCUCGUCCUGCUCGUCCUUCCACUAACAAGAA ....----...(((((.((((((((((((((...))))))))........)))))).(((----.-.((-------((..........))).)..)))................))))). ( -22.00) >DroYak_CAF1 35075 102 - 1 CUUC----CCCUUUUGAGUGUGUUGACCAACAAUGUUGGUCACAACAUGUACA--CACAG----C-CCA-------UCCCGUCGUCCUGGUCCUCCUGCUCGUCCUUCCACUAACAAGAA ....----...(((((.((((((((((((((...))))))))........)))--)))..----.-...-------....(.((..(.((....)).)..)).)..........))))). ( -21.60) >DroAna_CAF1 33907 108 - 1 UUCCGCCACCCCAUCGAGUGUGUUGACCGACAAUGUUGGUCACAACAUGCACACACACAGUCCGGUCCAGGGGUCACCCCG-----------AUUCCGGCAGUUGUUCC-CUAACAAGAU ....(((..........((((((((((((((...))))))))......)))))).........((....((((...)))).-----------...)))))..(((((..-..)))))... ( -32.90) >consensus CUCC____CCCUUUUGAGUGUGUUGACCAACAAUGUUGGUCACAACAUGUACA__CACAG____C_CCA_____UACCCCGUC______GUCCUUCUACUCGUCCUUCCACUAACAAGAA ..............(((((((((.(((((((...))))))))))...(((......))).....................................)))))).................. (-12.90 = -13.10 + 0.20)

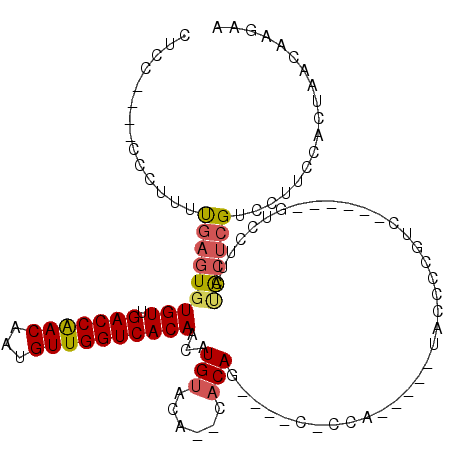
| Location | 8,604,371 – 8,604,478 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.20 |
| Mean single sequence MFE | -30.73 |
| Consensus MFE | -18.39 |
| Energy contribution | -18.45 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
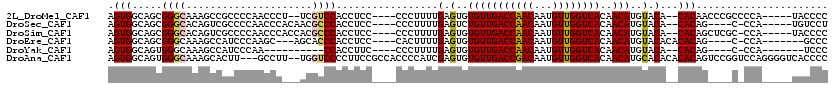
>2L_DroMel_CAF1 8604371 107 - 22407834 AGUGGCAGCGGGCAAAGCCGCCCCAACCCU--UCGUCCACCUCC----CCCUUUUGAGUGUGUUGACCAACAAUGUUGGUCACAACAUGUACA--CACAACCCGCCCCA-----UACCCC .((((..((((((......)))).......--............----.........((((((((((((((...))))))))........)))--))).....)).)))-----)..... ( -29.60) >DroSec_CAF1 32785 104 - 1 AGUGGCAGCGGGCACAGUCGCCCCAACCCACAACGCCCACCUCC----CCCUUUUGAGUCUGUUGACCAACAAUGUUGGUCACAACAUGUACA--CACAG----C-CCA-----UGUCCU .((((....((((......))))....)))).........(((.----.......)))..(((.(((((((...))))))))))(((((..(.--....)----.-.))-----)))... ( -27.60) >DroSim_CAF1 38472 108 - 1 AGUGGCAGCGGGCACAGUCGCCCCAACCCACCACGCCCACCUCC----CCCUUUUGAGUGUGUUGACCAACAAUGUUGGUCACAACAUGUACA--CACAGCUCGC-CCA-----UACCCC .(.((((((((((......)))).....................----.........((((((((((((((...))))))))........)))--))).))).))-)).-----...... ( -32.90) >DroEre_CAF1 34538 101 - 1 AGUGGCAGCGGGCAAAGCCAUCCCAAGC---AGCACCCACCUCC----CACUUUUGAGUGUGUUGACCAACAAUGUUGGUCACAACAUGUACACACACAG----C-CCA-------GCCC ...(((...((((...((........))---.............----......((.((((((((((((((...))))))))........)))))).)))----)-)).-------))). ( -29.60) >DroYak_CAF1 35115 92 - 1 AGUGGCAGUGGGCAAAGCCAUCCCAA----------CCACCUUC----CCCUUUUGAGUGUGUUGACCAACAAUGUUGGUCACAACAUGUACA--CACAG----C-CCA-------UCCC .(.(((.((((...............----------))))....----.........((((((((((((((...))))))))........)))--))).)----)-)).-------.... ( -25.96) >DroAna_CAF1 33935 115 - 1 AGUGGCAGUGGGCAAAGCACUU---GCCUU--UGGUCCCCUUCCGCCACCCCAUCGAGUGUGUUGACCGACAAUGUUGGUCACAACAUGCACACACACAGUCCGGUCCAGGGGUCACCCC .((((.((.(((((((((....---).)))--))..))))).)))).(((((((((.((((((((((((((...))))))))..........))))))....))))...)))))...... ( -38.70) >consensus AGUGGCAGCGGGCAAAGCCGCCCCAACCC___ACGCCCACCUCC____CCCUUUUGAGUGUGUUGACCAACAAUGUUGGUCACAACAUGUACA__CACAG____C_CCA_____UACCCC .(((.....((((.....................)))).................(.(..(((((((((((...))))))))..)))..).)...)))...................... (-18.39 = -18.45 + 0.06)
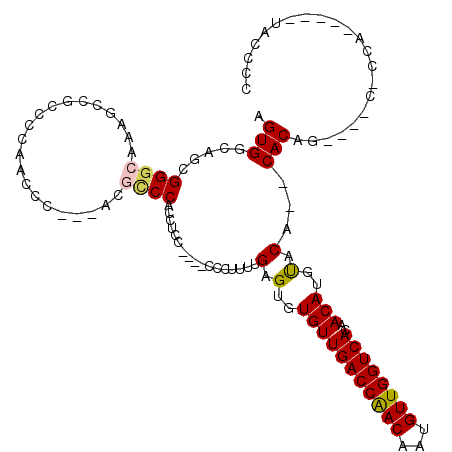
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:04 2006