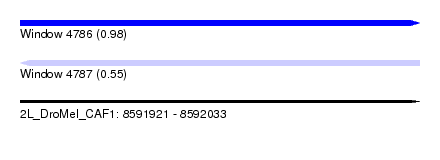
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,591,921 – 8,592,033 |
| Length | 112 |
| Max. P | 0.981261 |

| Location | 8,591,921 – 8,592,033 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.90 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -11.77 |
| Energy contribution | -13.63 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.37 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
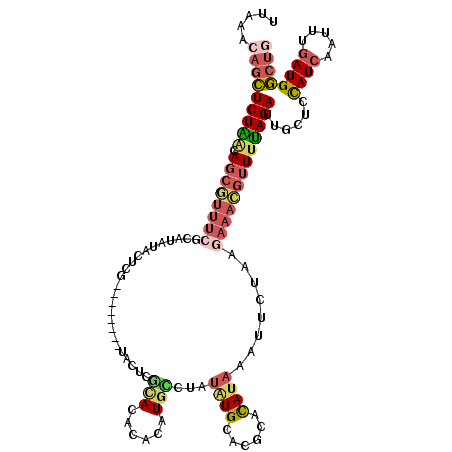
>2L_DroMel_CAF1 8591921 112 + 22407834 CAGCCAUCAAAUUGAUGGAGAAAUCUAAAACGUUUCUUAGAAUUUAUAUGCCUGCAUAUAGACAUGUGCGUACGAGGA--------CGAGUAUAUGCGAAACGCUGUUAGAGCUGUUUAA ((((((((.....))))......(((((..((((((......((((((((....))))))))(((((((...((....--------)).))))))).))))))...)))))))))..... ( -31.30) >DroSec_CAF1 27444 112 + 1 CAGCCAUCAAAUUGAUGGAGCGAUCUAAAACGUUUCUUAGAAUUUAUGUGCGUGCAUAUAGGCAUGUGUGUACGAGUA--------CGAGUAUAUGCGAAACGCUGUUAGAGCUGUUUAA ...(((((.....)))))(((..(((((..((((((.((..(((..((..((((((((((....))))))))))..))--------..)))...)).))))))...))))))))...... ( -33.80) >DroSim_CAF1 33052 112 + 1 CAGCCAUCAAAUUGAUGGAGCAAUCUAAAACGUUUCUUAGAAUUUAUGUGCGUGCAUAUAGGCAUGUGUGUACGAGUA--------CGAGUAUAUGCGAAACGCUGUUAGAGCUCUUUAA ....((((.....))))((((..(((((..((((((.((..(((..((..((((((((((....))))))))))..))--------..)))...)).))))))...)))))))))..... ( -33.80) >DroEre_CAF1 29130 120 + 1 CAGCCAUCAAAUUGAUAGACCCAUCUAAAACGUUUCUUAGAAUUUAUGUGCGUGCACAUAGACAUCUGUGUGUGUGUAUAUGGGAAAUGGGAAAUGGGAAAUGCUGUUAGAGCUGUUUAA ((((.(((.....))).......(((((..(((((((((...((((((((((..(((((((....)))))))..))))))))))...)))))))))((.....)).)))))))))..... ( -37.80) >DroYak_CAF1 29614 106 + 1 CAUCCAUCAAAUUGAUGGAGCCAUCUGAAACGUUUCUUAGAAUUUAUGUGCGUGCAUAUAGGCAUUUGUGUGCGAGUAUAUGGG--------------AAACGCUGCCAGAGCUGUUUAA ..((((((.....))))))((..((((..((((((((.......(((((((.(((((((((....))))))))).)))))))))--------------))))).)..))))))....... ( -32.71) >DroAna_CAF1 28464 93 + 1 CUGGCAUUAAAUUGAUAAUGCAAUCUAUACAA------GGACAUCGUA-----UCAUCACGGGAUGUGUAUGUGUGGGCA----------------UGGAAUGCUUUCAGACCUCUUUAA (((((((((......))))))...(((((((.------(.(((((.(.-----.......).))))).).)))))))(((----------------(...))))...))).......... ( -22.10) >consensus CAGCCAUCAAAUUGAUGGAGCAAUCUAAAACGUUUCUUAGAAUUUAUGUGCGUGCAUAUAGGCAUGUGUGUACGAGUA________CGAGUAUAUGCGAAACGCUGUUAGAGCUGUUUAA ...(((((.....))))).....(((((..((((((.......(((((((....)))))))(((....)))..........................))))))...)))))......... (-11.77 = -13.63 + 1.86)

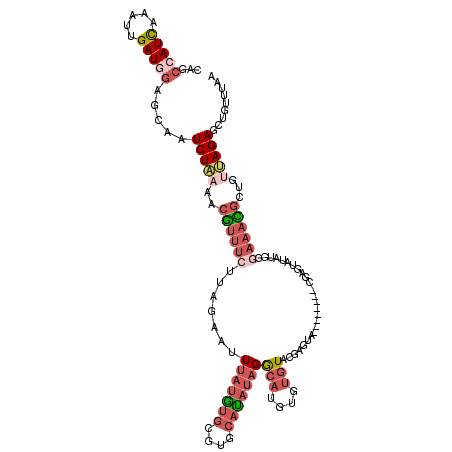
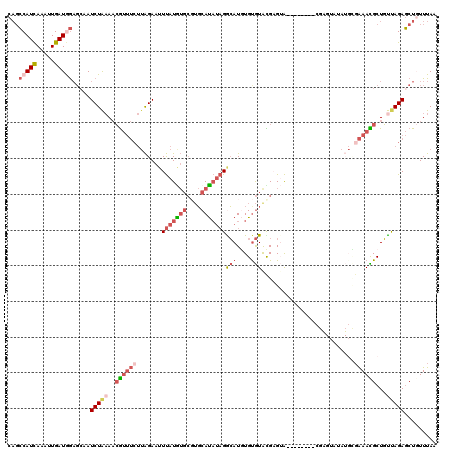
| Location | 8,591,921 – 8,592,033 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.90 |
| Mean single sequence MFE | -23.32 |
| Consensus MFE | -9.66 |
| Energy contribution | -10.70 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8591921 112 - 22407834 UUAAACAGCUCUAACAGCGUUUCGCAUAUACUCG--------UCCUCGUACGCACAUGUCUAUAUGCAGGCAUAUAAAUUCUAAGAAACGUUUUAGAUUUCUCCAUCAAUUUGAUGGCUG .....(((((((((.((((((((((...(((...--------.....))).))..((((((......))))))...........)))))))))))))......((((.....)))))))) ( -27.30) >DroSec_CAF1 27444 112 - 1 UUAAACAGCUCUAACAGCGUUUCGCAUAUACUCG--------UACUCGUACACACAUGCCUAUAUGCACGCACAUAAAUUCUAAGAAACGUUUUAGAUCGCUCCAUCAAUUUGAUGGCUG .....(((((((((.(((((((((((((((..((--------(............)))..))))))).......((.....)).)))))))))))))......((((.....)))))))) ( -25.20) >DroSim_CAF1 33052 112 - 1 UUAAAGAGCUCUAACAGCGUUUCGCAUAUACUCG--------UACUCGUACACACAUGCCUAUAUGCACGCACAUAAAUUCUAAGAAACGUUUUAGAUUGCUCCAUCAAUUUGAUGGCUG .....(((((((((.(((((((((((((((..((--------(............)))..))))))).......((.....)).)))))))))))))..))))((((.....)))).... ( -25.90) >DroEre_CAF1 29130 120 - 1 UUAAACAGCUCUAACAGCAUUUCCCAUUUCCCAUUUCCCAUAUACACACACACAGAUGUCUAUGUGCACGCACAUAAAUUCUAAGAAACGUUUUAGAUGGGUCUAUCAAUUUGAUGGCUG ..............((((..................((((....................((((((....))))))...(((((((....)))))))))))..((((.....)))))))) ( -21.10) >DroYak_CAF1 29614 106 - 1 UUAAACAGCUCUGGCAGCGUUU--------------CCCAUAUACUCGCACACAAAUGCCUAUAUGCACGCACAUAAAUUCUAAGAAACGUUUCAGAUGGCUCCAUCAAUUUGAUGGAUG .......(((((((.(((((((--------------(..........(((......)))...((((......))))........)))))))))))))..))((((((.....)))))).. ( -25.20) >DroAna_CAF1 28464 93 - 1 UUAAAGAGGUCUGAAAGCAUUCCA----------------UGCCCACACAUACACAUCCCGUGAUGA-----UACGAUGUCC------UUGUAUAGAUUGCAUUAUCAAUUUAAUGCCAG .......(((((....((((...)----------------))).....(((.(((.....))))))(-----(((((.....------)))))))))))((((((......))))))... ( -15.20) >consensus UUAAACAGCUCUAACAGCGUUUCGCAUAUACUCG________UACUCGCACACACAUGCCUAUAUGCACGCACAUAAAUUCUAAGAAACGUUUUAGAUUGCUCCAUCAAUUUGAUGGCUG .....(((((((((.((((((((........................(((......)))...((((......))))........)))))))))))))......((((.....)))))))) ( -9.66 = -10.70 + 1.04)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:02 2006