
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,574,946 – 8,575,039 |
| Length | 93 |
| Max. P | 0.887276 |

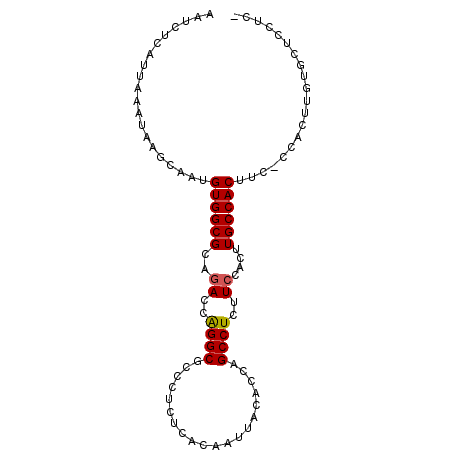
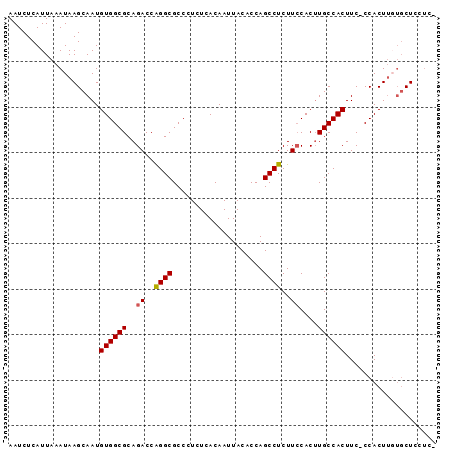
| Location | 8,574,946 – 8,575,039 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 95.04 |
| Mean single sequence MFE | -14.79 |
| Consensus MFE | -12.85 |
| Energy contribution | -12.89 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8574946 93 + 22407834 AAUCUCAUUAAAUAAGCAAUGUGGCGCAGACCAGGCGCCCUCUCACAAUUACACCAGCCUCUUCCACUUGCCACUUCCCCACUUGUGCUCCUC- ..............((((..((((((..((..((((....................))))..))....))))))...........))))....- ( -15.17) >DroSec_CAF1 10600 92 + 1 AAUCUCAUUAAAUAAGCAAUGUGGCGCAGACCAGGCGCCCUCUCACAAUUACACCAGCCUCUUCCACUUGCCACUUC-CCACUUGUGCUCCUC- ...........(((((....((((((..((..((((....................))))..))....))))))...-...))))).......- ( -15.45) >DroSim_CAF1 15924 92 + 1 AAUCUCAUUAAAUAAGCAAUGUGGCGCAGACCAGGCGCCCUCUCACAAUUACACCAGCCUCUUCCACUUGCCACUUC-CCACUUGUGCUCCUC- ...........(((((....((((((..((..((((....................))))..))....))))))...-...))))).......- ( -15.45) >DroEre_CAF1 10900 92 + 1 AAUCUCAUUAAAUAAGCAAUGUGGCGCAGACCGGGCGCCCUCUCACAAUUACACCAGCCUCUUGCACAUGCCACUUC-CCACUUUUUCUCCUC- ....................(((((((((...((((....................)))).))))....)))))...-...............- ( -14.35) >DroYak_CAF1 11634 93 + 1 AAUCUCAUUAAAUAAGCAAUGUGGCGCAGACCAGGCGCCCUCUUACAAUUACACCAGCCUCUUCCACUUGCCACUUC-CCACUGUAGCUCCACC ....................((((((..((..((((....................))))..))....))))))...-................ ( -13.55) >consensus AAUCUCAUUAAAUAAGCAAUGUGGCGCAGACCAGGCGCCCUCUCACAAUUACACCAGCCUCUUCCACUUGCCACUUC_CCACUUGUGCUCCUC_ ....................((((((..((..((((....................))))..))....)))))).................... (-12.85 = -12.89 + 0.04)

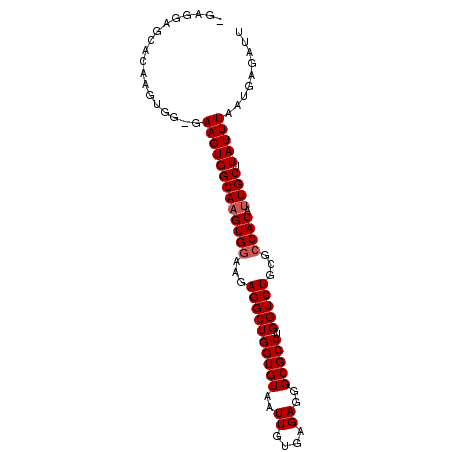
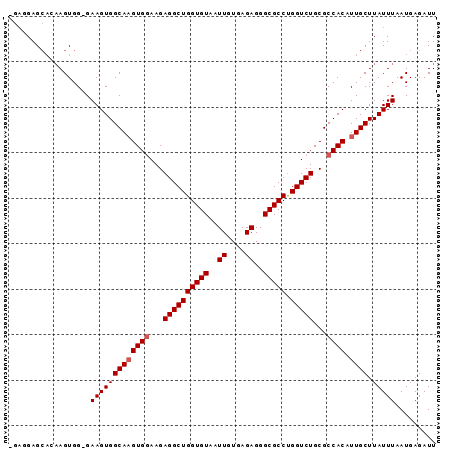
| Location | 8,574,946 – 8,575,039 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 95.04 |
| Mean single sequence MFE | -25.34 |
| Consensus MFE | -22.94 |
| Energy contribution | -23.34 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.576111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8574946 93 - 22407834 -GAGGAGCACAAGUGGGGAAGUGGCAAGUGGAAGAGGCUGGUGUAAUUGUGAGAGGGCGCCUGGUCUGCGCCACAUUGCUUAUUUAAUGAGAUU -.................(((((((((((((...((((((((((..((....))..))))).)))))...)))).)))).)))))......... ( -25.00) >DroSec_CAF1 10600 92 - 1 -GAGGAGCACAAGUGG-GAAGUGGCAAGUGGAAGAGGCUGGUGUAAUUGUGAGAGGGCGCCUGGUCUGCGCCACAUUGCUUAUUUAAUGAGAUU -...............-.(((((((((((((...((((((((((..((....))..))))).)))))...)))).)))).)))))......... ( -25.00) >DroSim_CAF1 15924 92 - 1 -GAGGAGCACAAGUGG-GAAGUGGCAAGUGGAAGAGGCUGGUGUAAUUGUGAGAGGGCGCCUGGUCUGCGCCACAUUGCUUAUUUAAUGAGAUU -...............-.(((((((((((((...((((((((((..((....))..))))).)))))...)))).)))).)))))......... ( -25.00) >DroEre_CAF1 10900 92 - 1 -GAGGAGAAAAAGUGG-GAAGUGGCAUGUGCAAGAGGCUGGUGUAAUUGUGAGAGGGCGCCCGGUCUGCGCCACAUUGCUUAUUUAAUGAGAUU -.....(((.(((..(-...(((((....(((...(((((((((..((....))..))).)))))))))))))).)..))).)))......... ( -24.20) >DroYak_CAF1 11634 93 - 1 GGUGGAGCUACAGUGG-GAAGUGGCAAGUGGAAGAGGCUGGUGUAAUUGUAAGAGGGCGCCUGGUCUGCGCCACAUUGCUUAUUUAAUGAGAUU ((..(.(((((.....-...)))))..((((...((((((((((..((....))..))))).)))))...)))).)..)).............. ( -27.50) >consensus _GAGGAGCACAAGUGG_GAAGUGGCAAGUGGAAGAGGCUGGUGUAAUUGUGAGAGGGCGCCUGGUCUGCGCCACAUUGCUUAUUUAAUGAGAUU ..................(((((((((((((...((((((((((..((....))..))))).)))))...)))).)))).)))))......... (-22.94 = -23.34 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:56 2006