
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,560,998 – 8,561,161 |
| Length | 163 |
| Max. P | 0.999823 |

| Location | 8,560,998 – 8,561,095 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 92.06 |
| Mean single sequence MFE | -33.13 |
| Consensus MFE | -25.65 |
| Energy contribution | -26.15 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.712572 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8560998 97 + 22407834 UUA----UAUAUUAAGAGCUGAUUUUUU-CUCUCAACUCUUGUGA---UUGCAAUGGCGGCGCGGCCAUUUUGCAAAAGGCUGCAACAAAAUGGCGGCCGCAAAA- ...----.....((((((.(((......-...))).))))))...---........(((((.((.((((((((((......))))...)))))))))))))....- ( -28.60) >DroSec_CAF1 70651 102 + 1 UUAUUUGUAUAUUAAGAGCUGAUUUUUUGCUCUCAACUCUUGUGA---UUGCAAUGGCGGCGCGGCCAUUUUGCAAAAGGCUGCAACAAAAUGGCUGCCGCAAAA- ....(((((...((((((.(((..........))).))))))...---.)))))..(((((..((((((((((((......))))...)))))))))))))....- ( -33.00) >DroSim_CAF1 74653 102 + 1 UUAUUUGUAUAUUAAGAGCUGAUUUUUUGCUCUCAACUCUUGUGA---UUGCAAUGGCGGCGCGGCCAUUUUGCAAAAGGCUGCAACAAAAUGGCUGCCGCAAAA- ....(((((...((((((.(((..........))).))))))...---.)))))..(((((..((((((((((((......))))...)))))))))))))....- ( -33.00) >DroEre_CAF1 72225 102 + 1 UUAUUUGUAUAUUAAGAGCUGAUUUUUUGCUCUCAACUCUUGUGC---UCGCAAUGGCGGCGCGGCCAUUUUGCAAAAGGCUGCAACAAAAUGGCGGCCGCAAAA- ...(((((....((((((.(((..........))).)))))).((---.(((....)))))((.(((((((((((......))))...))))))).)).))))).- ( -30.80) >DroYak_CAF1 71875 102 + 1 UUAUUUGUAUAUUAAGAGCUGAUUUUUUGCUCCCAACUCUUGUGU---UCGCAAUGGCGGCGCGGCCAUUUUGCAAAAGGCUGCAACAAAAUGGCGGCCGCAAAA- ....((((....((((((.((............)).))))))...---..))))..(((((.((.((((((((((......))))...)))))))))))))....- ( -29.00) >DroAna_CAF1 55208 102 + 1 UUAUUUGUAUAUUAAGAGCUGAUUUUUUGCACUC-ACUGUUGUGAGUGUGGCUAUGGCGGC--CGCCAUUUUGCAAA-UGCUGCAACAAAAUGGCGGCCGCAAAAA ................((((........((((((-((....))))))))))))...(((((--((((((((((((..-...))))...)))))))))))))..... ( -44.40) >consensus UUAUUUGUAUAUUAAGAGCUGAUUUUUUGCUCUCAACUCUUGUGA___UUGCAAUGGCGGCGCGGCCAUUUUGCAAAAGGCUGCAACAAAAUGGCGGCCGCAAAA_ ............((((((.(((..........))).))))))..............(((((...(((((((((((......))))...))))))).)))))..... (-25.65 = -26.15 + 0.50)

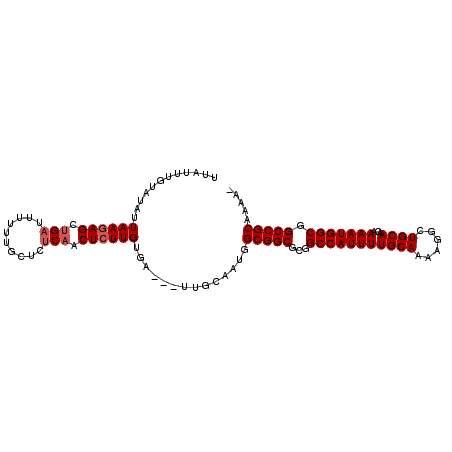
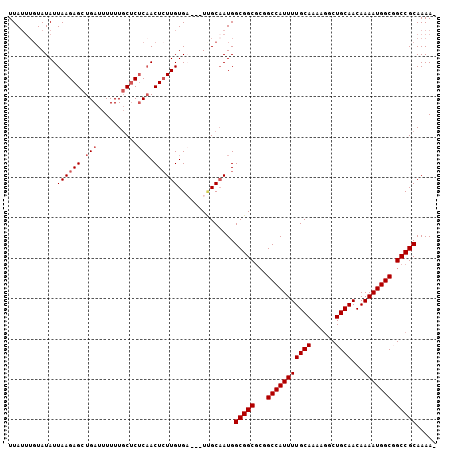
| Location | 8,560,998 – 8,561,095 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 92.06 |
| Mean single sequence MFE | -33.97 |
| Consensus MFE | -30.62 |
| Energy contribution | -30.82 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.04 |
| Mean z-score | -4.15 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.17 |
| SVM RNA-class probability | 0.999823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8560998 97 - 22407834 -UUUUGCGGCCGCCAUUUUGUUGCAGCCUUUUGCAAAAUGGCCGCGCCGCCAUUGCAA---UCACAAGAGUUGAGAG-AAAAAAUCAGCUCUUAAUAUA----UAA -....((((((((((((((((...........))))))))).)).)))))........---....(((((((((...-......)))))))))......----... ( -33.10) >DroSec_CAF1 70651 102 - 1 -UUUUGCGGCAGCCAUUUUGUUGCAGCCUUUUGCAAAAUGGCCGCGCCGCCAUUGCAA---UCACAAGAGUUGAGAGCAAAAAAUCAGCUCUUAAUAUACAAAUAA -....(((((.((((((((((...........))))))))))...)))))........---....(((((((((..........)))))))))............. ( -32.20) >DroSim_CAF1 74653 102 - 1 -UUUUGCGGCAGCCAUUUUGUUGCAGCCUUUUGCAAAAUGGCCGCGCCGCCAUUGCAA---UCACAAGAGUUGAGAGCAAAAAAUCAGCUCUUAAUAUACAAAUAA -....(((((.((((((((((...........))))))))))...)))))........---....(((((((((..........)))))))))............. ( -32.20) >DroEre_CAF1 72225 102 - 1 -UUUUGCGGCCGCCAUUUUGUUGCAGCCUUUUGCAAAAUGGCCGCGCCGCCAUUGCGA---GCACAAGAGUUGAGAGCAAAAAAUCAGCUCUUAAUAUACAAAUAA -(((((..((.((((((((((...........)))))))))).))(((((....))).---)).)))))(((((((((.........))))))))).......... ( -33.70) >DroYak_CAF1 71875 102 - 1 -UUUUGCGGCCGCCAUUUUGUUGCAGCCUUUUGCAAAAUGGCCGCGCCGCCAUUGCGA---ACACAAGAGUUGGGAGCAAAAAAUCAGCUCUUAAUAUACAAAUAA -....((((((((((((((((...........))))))))).)).)))))........---........(((((((((.........))))))))).......... ( -32.20) >DroAna_CAF1 55208 102 - 1 UUUUUGCGGCCGCCAUUUUGUUGCAGCA-UUUGCAAAAUGGCG--GCCGCCAUAGCCACACUCACAACAGU-GAGUGCAAAAAAUCAGCUCUUAAUAUACAAAUAA .....((((((((((((((((.......-...)))))))))))--)))))........(((((((....))-)))))............................. ( -40.40) >consensus _UUUUGCGGCCGCCAUUUUGUUGCAGCCUUUUGCAAAAUGGCCGCGCCGCCAUUGCAA___UCACAAGAGUUGAGAGCAAAAAAUCAGCUCUUAAUAUACAAAUAA .....(((((.((((((((((...........))))))))))...)))))...............(((((((((..........)))))))))............. (-30.62 = -30.82 + 0.20)

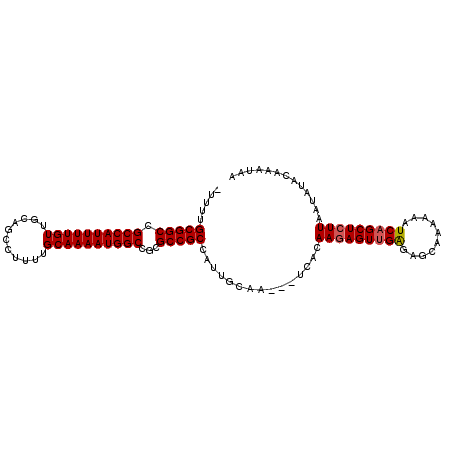
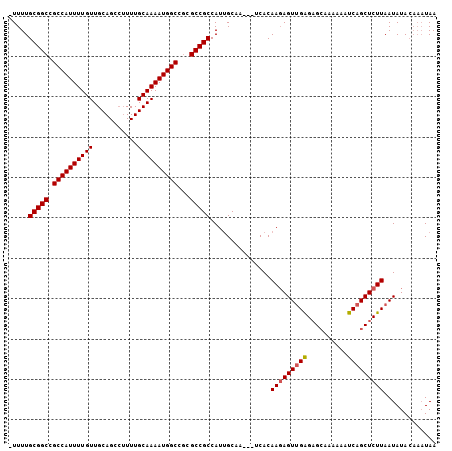
| Location | 8,561,024 – 8,561,131 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.13 |
| Mean single sequence MFE | -34.82 |
| Consensus MFE | -23.88 |
| Energy contribution | -24.13 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8561024 107 - 22407834 CACUCACUUAACACUCAUC----AGGCGAUGCAAUUUGUU-UUUUGCGGCCGCCAUUUUGUUGCAGCCUUUUGCAAAAUGGCCGCG--CCGCCAUUGC---AA---UCACAAGAGUUGAG ......((((((.((....----..((((((.........-....((((((((((((((((...........))))))))).)).)--))))))))))---..---.....)).)))))) ( -32.93) >DroSec_CAF1 70682 107 - 1 CACUCACUUAACACUCAUC----AGGCGAUGCAAUUGGUU-UUUUGCGGCAGCCAUUUUGUUGCAGCCUUUUGCAAAAUGGCCGCG--CCGCCAUUGC---AA---UCACAAGAGUUGAG ..((((......((((...----.((((.(((((......-..)))))((.((((((((((...........)))))))))).)).--.)))).(((.---..---...))))))))))) ( -32.80) >DroEre_CAF1 72256 107 - 1 CACUCACUUAACACUCAUC----AGGCGAUGCAAUUUGUU-UUUUGCGGCCGCCAUUUUGUUGCAGCCUUUUGCAAAAUGGCCGCG--CCGCCAUUGC---GA---GCACAAGAGUUGAG ......((((((.......----.((((.(((((......-..)))))..)))).((((((.((.(((...........))).))(--((((....))---).---)))))))))))))) ( -33.90) >DroWil_CAF1 59916 102 - 1 CAUCCACUUAACACUCAUC----AGGCGAUGCAAUUUGU----UUGUGGCCGCCAUUUUGUUGCAG---UUUGCAAAAUGGCUGCCGUUUGCCAUUGCCUCGA---AUACAACAGA---- .................((----(((((((((((.....----...((((.((((((((((.....---...)))))))))).)))).))).)))))))).))---..........---- ( -32.60) >DroYak_CAF1 71906 107 - 1 CACUCACUUAACACUCAUC----AGGCGAUGCAAUUUGUU-UUUUGCGGCCGCCAUUUUGUUGCAGCCUUUUGCAAAAUGGCCGCG--CCGCCAUUGC---GA---ACACAAGAGUUGGG ......((((((...((((----....))))...(((((.-....((((((((((((((((...........))))))))).)).)--))))....))---))---).......)))))) ( -32.00) >DroAna_CAF1 55239 111 - 1 CACUCACUUAACACUCAUCGGGCAGGCAGUGCAAUUUAUUUUUUUGCGGCCGCCAUUUUGUUGCAGCA-UUUGCAAAAUGGCG--G--CCGCCAUAGC---CACACUCACAACAGU-GAG ..((((((............(((.(((...((((.........))))((((((((((((((.......-...)))))))))))--)--)))))...))---)...........)))-))) ( -44.70) >consensus CACUCACUUAACACUCAUC____AGGCGAUGCAAUUUGUU_UUUUGCGGCCGCCAUUUUGUUGCAGCCUUUUGCAAAAUGGCCGCG__CCGCCAUUGC___GA___UCACAAGAGUUGAG ........................((((.(((((.........)))))((.((((((((((...........)))))))))).))....))))........................... (-23.88 = -24.13 + 0.25)


| Location | 8,561,056 – 8,561,161 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 76.88 |
| Mean single sequence MFE | -17.87 |
| Consensus MFE | -13.07 |
| Energy contribution | -12.93 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8561056 105 - 22407834 CCAUC-AAACACCCACUCAACGCACACCACC--CACUCACUUAACACUCAUC----AGGCGAUGCAAUUUGUU-UUUUGCGGCCGCCAUUUUGUUGCAGCCUUUUGCAAAAUG .....-.........................--...................----.((((.(((((......-..)))))..))))......((((((....)))))).... ( -17.10) >DroSec_CAF1 70714 100 - 1 ------CAACACCCACUCAACACACACCACC--CACUCACUUAACACUCAUC----AGGCGAUGCAAUUGGUU-UUUUGCGGCAGCCAUUUUGUUGCAGCCUUUUGCAAAAUG ------.........................--...................----......(((((..(((.-...((((((((.....)))))))))))..)))))..... ( -17.40) >DroSim_CAF1 74716 82 - 1 ------CAACACCC--------------------ACUCACUUAACACUCAUC----AGGCGAUGCAAUUUGUU-UUUUGCGGCAGCCAUUUUGUUGCAGCCUUUUGCAAAAUG ------........--------------------..............((..----((((..(((((......-..)))))(((((......))))).))))..))....... ( -16.60) >DroEre_CAF1 72288 105 - 1 CCACC-CAACACACACUCAACACACACCACC--CACUCACUUAACACUCAUC----AGGCGAUGCAAUUUGUU-UUUUGCGGCCGCCAUUUUGUUGCAGCCUUUUGCAAAAUG .....-.........................--...................----.((((.(((((......-..)))))..))))......((((((....)))))).... ( -17.10) >DroWil_CAF1 59949 97 - 1 CCUCU-G----GCCCCGCAUCCGCCGCCAGCAACAUCCACUUAACACUCAUC----AGGCGAUGCAAUUUGU----UUGUGGCCGCCAUUUUGUUGCAG---UUUGCAAAAUG ...((-(----((...((....)).)))))......................----.((((.(((((.....----)))))..))))(((((((.....---...))))))). ( -22.40) >DroAna_CAF1 55271 101 - 1 CCAUCACAUCACCUCCUCAC---------CC--CACUCACUUAACACUCAUCGGGCAGGCAGUGCAAUUUAUUUUUUUGCGGCCGCCAUUUUGUUGCAGCA-UUUGCAAAAUG ....................---------..--....................(((.(((...((((.........)))).))))))......((((((..-.)))))).... ( -16.60) >consensus CCA_C_CAACACCCACUCAAC_CACACCACC__CACUCACUUAACACUCAUC____AGGCGAUGCAAUUUGUU_UUUUGCGGCCGCCAUUUUGUUGCAGCCUUUUGCAAAAUG .........................................................(((..(((((.........)))))...)))......((((((....)))))).... (-13.07 = -12.93 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:49 2006