
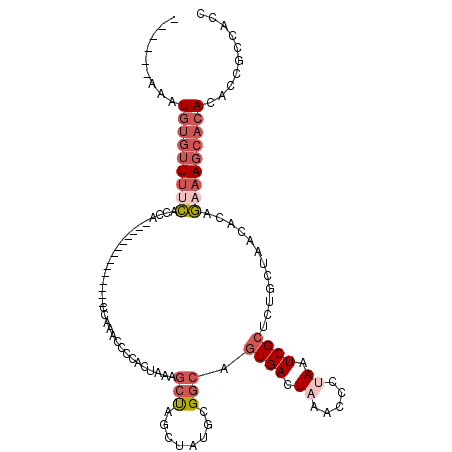
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 127,917 – 128,012 |
| Length | 95 |
| Max. P | 0.569321 |

| Location | 127,917 – 128,012 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 76.48 |
| Mean single sequence MFE | -19.06 |
| Consensus MFE | -8.18 |
| Energy contribution | -9.77 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 127917 95 + 22407834 ------AAAUGUGUUUUCACCA--------------CCACACCCGACUAAAGCUAGCUAU-UGGCAGUGACUAAAUCCUAAUCGCUCUGCUAACACAGAAAGCACAUACCGCCGCC ------..((((((((((....--------------........(.(((....))))..(-(((((((((.((.....)).))))..))))))....))))))))))......... ( -19.30) >DroPse_CAF1 15360 112 + 1 GAUAGCAAUUUAUGUGUU-CCGAAGGGAGUUUAACCCCAAACC-UUCUUU-CAUAUCUAUCCGCCUAUGAAUAAAACCUAAUCGCUGUGCUAACACAACAAGCACACACUCGUAC- (((((......(((((..-..(((((...............))-)))...-))))))))))......................(.((((((.........)))))))........- ( -16.96) >DroSec_CAF1 10465 96 + 1 ------AAAUGUGUUUUCACCA--------------CCAAACCCCACUAAAGCUAGAUAUGCGGCAGUGACUAAUCCCAAAUCGCUCUGCUAACACAGAAAGCACACACCGCCGCC ------...(((((((((....--------------..........(((....)))....((((.(((((...........))))))))).......))))))))).......... ( -19.80) >DroSim_CAF1 10185 96 + 1 ------AAAUGUGUUUUCACCA--------------CCAAACCCCACUAAAGCUAGAUAUGCGGCAGUGACUAAUCCCAAAUCGCUCUGCUAACACAGAAAGCACACACCGCCGCC ------...(((((((((....--------------..........(((....)))....((((.(((((...........))))))))).......))))))))).......... ( -19.80) >DroEre_CAF1 10449 96 + 1 ------AAAUGUGUUUUCUCCA--------------CCAUACCGCACUAUAGCUAGCUAUGCGGCUGUGACUAAACCCUAAUCGCUCUGCUAACACUGAAAGCACACACCGCCACC ------...(((((((((....--------------.(((((((((...(((....)))))))).))))..............((...)).......))))))))).......... ( -21.30) >DroYak_CAF1 10265 95 + 1 ------AAAUGUGUUUCCUCCA--------------CCAAACC-CACUAUAGCCAGUUAUGCGGCUGUGACUAAACCCUAAUCGCUCUGCUAACACAGAAAGCACACACCGCCACC ------...(((((((......--------------.......-...(((((((........)))))))................((((......))))))))))).......... ( -17.20) >consensus ______AAAUGUGUUUUCACCA______________CCAAACCCCACUAAAGCUAGCUAUGCGGCAGUGACUAAACCCUAAUCGCUCUGCUAACACAGAAAGCACACACCGCCACC .........(((((((((.................................(((........))).((((.((.....)).))))............))))))))).......... ( -8.18 = -9.77 + 1.59)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:23:42 2006