| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,559,320 – 8,559,421 |
| Length | 101 |
| Max. P | 0.823029 |

| Location | 8,559,320 – 8,559,421 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.10 |
| Mean single sequence MFE | -24.97 |
| Consensus MFE | -17.50 |
| Energy contribution | -19.20 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
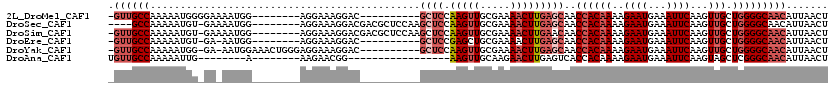
>2L_DroMel_CAF1 8559320 101 + 22407834 -GUUGCCAAAAAUGGGGAAAAUGG--------AGGAAAGGAC----------GCUCCAAGUUGCGAAAACUUGAGCAACCACAAAAGAAUGAAAUUCAAGUUGCUGGGGCAACAUUAACU -((((((.................--------..........----------((((.(((((.....)))))))))..((((((..((((...))))...))).)))))))))....... ( -25.70) >DroSec_CAF1 68999 107 + 1 ----GCCAAAAAUGU-GAAAAUGG--------AGGAAAGGACGACGCUCCAAGCUCCAAGUUGCGAAAACUUGAGCAACCACAAAAGAAUGAAAUUCAAGUUGCUGGGGCAACAUUAACU ----........(((-(....(((--------((............))))).((((.(((((.....)))))))))...))))...((((...))))..(((((....)))))....... ( -25.80) >DroSim_CAF1 70313 110 + 1 -GUUGCCAAAAAUGU-GAAAAUGG--------AGGAAAGGACGACGCUCCAAGCUCCAAGUUGCGAAAACUUGAACAACCACAAAAGAAUGAAAUUCAAGUUGCUGGGGCAACAUUAACU -((((((.....(((-.....(((--------((............))))).....((((((.....)))))).))).((((((..((((...))))...))).)))))))))....... ( -27.00) >DroEre_CAF1 70589 99 + 1 -GUUGCCAAAAAUGU-GA-AAUGG--------AGGAAAGGAC----------GCUCCGAGCUGCGAAAACUUGAGCAACCACAAAAGAAUGAAAUUCAAGUUGCUGGGGCAACAUUAACU -((((((........-..-..(((--------(((......)----------.))))).(((.(((....))))))..((((((..((((...))))...))).)))))))))....... ( -23.80) >DroYak_CAF1 70202 107 + 1 -GUUGCCAAAAAUGG-GA-AAUGGAAACUGGGAGGAAAGGAC----------GCUCCAAGUUGCGAAAACUUGAGCAACCACAAAAGAAUGAAAUUCAAGUUGCUGGGGCAACAUUAACU -((((((...(((..-((-(..(....)..........((..----------((((.(((((.....)))))))))..))..............)))..))).....))))))....... ( -27.10) >DroAna_CAF1 53743 87 + 1 UGUUGCCAAAAAUUG--------A--------AAGAACGG-----------------AAGUUGCAAGAACUUGAGUCACCACAAAAGAAUGAAAUUCAAGUAGCUCGGGCAACAUUAACU (((((((........--------.--------...(((..-----------------..)))((....((((((((...((........))..)))))))).))...)))))))...... ( -20.40) >consensus _GUUGCCAAAAAUGG_GA_AAUGG________AGGAAAGGAC__________GCUCCAAGUUGCGAAAACUUGAGCAACCACAAAAGAAUGAAAUUCAAGUUGCUGGGGCAACAUUAACU .((((((.............................................((((.(((((.....)))))))))..((((((..((((...))))...))).)))))))))....... (-17.50 = -19.20 + 1.70)


| Location | 8,559,320 – 8,559,421 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.10 |
| Mean single sequence MFE | -23.55 |
| Consensus MFE | -15.54 |
| Energy contribution | -15.98 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
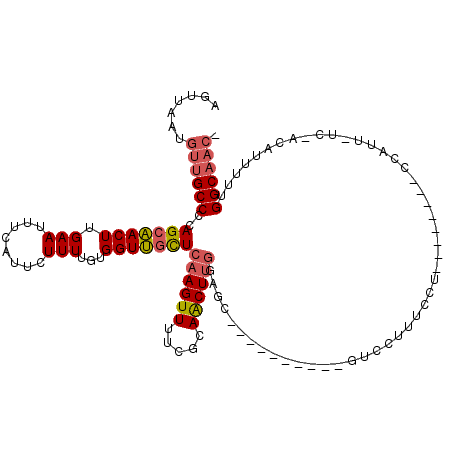
>2L_DroMel_CAF1 8559320 101 - 22407834 AGUUAAUGUUGCCCCAGCAACUUGAAUUUCAUUCUUUUGUGGUUGCUCAAGUUUUCGCAACUUGGAGC----------GUCCUUUCCU--------CCAUUUUCCCCAUUUUUGGCAAC- .......((((((...(((((((((...((((......))))....)))))))...))....(((((.----------(......)))--------)))..............))))))- ( -23.10) >DroSec_CAF1 68999 107 - 1 AGUUAAUGUUGCCCCAGCAACUUGAAUUUCAUUCUUUUGUGGUUGCUCAAGUUUUCGCAACUUGGAGCUUGGAGCGUCGUCCUUUCCU--------CCAUUUUC-ACAUUUUUGGC---- ..((((.(((((....)))))))))............(((((((((..........))))).(((((...(((......)))....))--------)))....)-)))........---- ( -22.90) >DroSim_CAF1 70313 110 - 1 AGUUAAUGUUGCCCCAGCAACUUGAAUUUCAUUCUUUUGUGGUUGUUCAAGUUUUCGCAACUUGGAGCUUGGAGCGUCGUCCUUUCCU--------CCAUUUUC-ACAUUUUUGGCAAC- .......((((((...(((((((((((.((((......))))..)))))))))...))....(((((...(((......)))....))--------))).....-........))))))- ( -29.00) >DroEre_CAF1 70589 99 - 1 AGUUAAUGUUGCCCCAGCAACUUGAAUUUCAUUCUUUUGUGGUUGCUCAAGUUUUCGCAGCUCGGAGC----------GUCCUUUCCU--------CCAUU-UC-ACAUUUUUGGCAAC- .......((((((...(((((((((...((((......))))....)))))))...)).....((((.----------(......)))--------))...-..-........))))))- ( -22.70) >DroYak_CAF1 70202 107 - 1 AGUUAAUGUUGCCCCAGCAACUUGAAUUUCAUUCUUUUGUGGUUGCUCAAGUUUUCGCAACUUGGAGC----------GUCCUUUCCUCCCAGUUUCCAUU-UC-CCAUUUUUGGCAAC- .......((((((...(((((((((...((((......))))....)))))))...))((((.((((.----------(......))))).))))......-..-........))))))- ( -24.70) >DroAna_CAF1 53743 87 - 1 AGUUAAUGUUGCCCGAGCUACUUGAAUUUCAUUCUUUUGUGGUGACUCAAGUUCUUGCAACUU-----------------CCGUUCUU--------U--------CAAUUUUUGGCAACA ......(((((((((((..((((((...((((......))))....)))))).)))).(((..-----------------..)))...--------.--------........))))))) ( -18.90) >consensus AGUUAAUGUUGCCCCAGCAACUUGAAUUUCAUUCUUUUGUGGUUGCUCAAGUUUUCGCAACUUGGAGC__________GUCCUUUCCU________CCAUU_UC_ACAUUUUUGGCAAC_ .......((((((..(((((((.(((........)))...)))))))((((((.....)))))).................................................)))))). (-15.54 = -15.98 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:44 2006