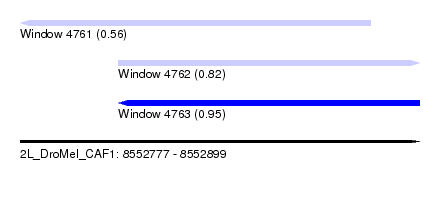
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,552,777 – 8,552,899 |
| Length | 122 |
| Max. P | 0.953885 |

| Location | 8,552,777 – 8,552,884 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 89.64 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -19.31 |
| Energy contribution | -20.88 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8552777 107 - 22407834 CAUUCAUUCCCAUC--CAUCCCCUUUGGCUAGGAACCUCGUGUCGCCAGCGGGAUUGUCAACGCCGUUGACCUCAUCUCCACCGUCCAACUGCAAUAAGCCUCGAAUGG (((((.........--((..(((.(((((((.(.....).))..))))).)))..)).....((.((((((............)).)))).))..........))))). ( -22.80) >DroSec_CAF1 62420 106 - 1 CAUUCAUUCCCGUC--CAACCCCU-UGGCCAGGAACCUCCUGUCGCCACCGGGAUUGUCAACGCCGUUGACCUCAUCUCCACCGUCCAACUGCAAUAAGCCUCGAAUGG (((((.........--((((((..-((((((((.....))))..))))..))).))).....((.((((((............)).)))).))..........))))). ( -27.40) >DroSim_CAF1 63342 106 - 1 CAUUCAUUCCCAUC--CAUCCCCU-UGGCCAGAAACCUCCUGUCGCCAGCGGGAUUGUCAACGCCGUUGACCUCAUCUCCACCGUCCAACUGCAAUAAGCCUCGAAUGG (((((.........--.(((((.(-(((((((.......)))..))))).))))).......((.((((((............)).)))).))..........))))). ( -25.70) >DroYak_CAF1 63593 109 - 1 CAUUCAUUCAUUCCCACAUCCCCUUUGGACAGAAACCUCCAGUCGCCAGUGGGGUUGUCAACGCCGUUGACCUCAUCUCCACUGUCCAAUGGCUUUAAGCCUCGAAUGG ....(((((...............((((((((.((((.(((.(....).)))))))((((((...))))))..........)))))))).(((.....)))..))))). ( -33.30) >consensus CAUUCAUUCCCAUC__CAUCCCCU_UGGCCAGAAACCUCCUGUCGCCAGCGGGAUUGUCAACGCCGUUGACCUCAUCUCCACCGUCCAACUGCAAUAAGCCUCGAAUGG (((((...............(((.(((((((((.....))))..))))).)))...((((((...))))))................................))))). (-19.31 = -20.88 + 1.56)

| Location | 8,552,807 – 8,552,899 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 89.05 |
| Mean single sequence MFE | -31.44 |
| Consensus MFE | -24.61 |
| Energy contribution | -25.92 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8552807 92 + 22407834 GAGAUGAGGUCAACGGCGUUGACAAUCCCGCUGGCGACACGAGGUUCCUAGCCAAAGGGGAUG--GAUGGGAAUGAAUGGAUGGGGACAUCCAU ........(((..(((((..........)))))..))).....(((((((.(((.......))--).)))))))..(((((((....))))))) ( -32.10) >DroSec_CAF1 62450 91 + 1 GAGAUGAGGUCAACGGCGUUGACAAUCCCGGUGGCGACAGGAGGUUCCUGGCCA-AGGGGUUG--GACGGGAAUGAAUGGAUGGGGACAUCCAU .........(((....((((..(((((((..((((..((((.....))))))))-.)))))))--))))....)))(((((((....))))))) ( -37.60) >DroSim_CAF1 63372 91 + 1 GAGAUGAGGUCAACGGCGUUGACAAUCCCGCUGGCGACAGGAGGUUUCUGGCCA-AGGGGAUG--GAUGGGAAUGAAUGGAUAGGGACAUCCAU ........((((((...)))))).(((((..((((..(((((...)))))))))-..)))))(--((((..................))))).. ( -27.47) >DroYak_CAF1 63623 94 + 1 GAGAUGAGGUCAACGGCGUUGACAACCCCACUGGCGACUGGAGGUUUCUGUCCAAAGGGGAUGUGGGAAUGAAUGAAUGGAUGGGGACAUCCCC ........((((((...))))))...(((((.((.(((.....))).))((((.....)))))))))...........(((((....))))).. ( -28.60) >consensus GAGAUGAGGUCAACGGCGUUGACAAUCCCGCUGGCGACAGGAGGUUCCUGGCCA_AGGGGAUG__GAUGGGAAUGAAUGGAUGGGGACAUCCAU ........((((((...))))))..((((..((((..(((((...)))))))))..))))................(((((((....))))))) (-24.61 = -25.92 + 1.31)

| Location | 8,552,807 – 8,552,899 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 89.05 |
| Mean single sequence MFE | -25.78 |
| Consensus MFE | -20.09 |
| Energy contribution | -21.52 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.953885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8552807 92 - 22407834 AUGGAUGUCCCCAUCCAUUCAUUCCCAUC--CAUCCCCUUUGGCUAGGAACCUCGUGUCGCCAGCGGGAUUGUCAACGCCGUUGACCUCAUCUC (((((((....)))))))...........--.(((((..(((((((.(.....).))..))))).))))).((((((...))))))........ ( -23.60) >DroSec_CAF1 62450 91 - 1 AUGGAUGUCCCCAUCCAUUCAUUCCCGUC--CAACCCCU-UGGCCAGGAACCUCCUGUCGCCACCGGGAUUGUCAACGCCGUUGACCUCAUCUC (((((((....)))))))...........--...(((..-((((((((.....))))..))))..)))...((((((...))))))........ ( -28.30) >DroSim_CAF1 63372 91 - 1 AUGGAUGUCCCUAUCCAUUCAUUCCCAUC--CAUCCCCU-UGGCCAGAAACCUCCUGUCGCCAGCGGGAUUGUCAACGCCGUUGACCUCAUCUC (((((((....)))))))...........--.(((((.(-(((((((.......)))..))))).))))).((((((...))))))........ ( -25.50) >DroYak_CAF1 63623 94 - 1 GGGGAUGUCCCCAUCCAUUCAUUCAUUCCCACAUCCCCUUUGGACAGAAACCUCCAGUCGCCAGUGGGGUUGUCAACGCCGUUGACCUCAUCUC ((((....))))...............(((((.......(((((........)))))......)))))(..((((((...))))))..)..... ( -25.72) >consensus AUGGAUGUCCCCAUCCAUUCAUUCCCAUC__CAUCCCCU_UGGCCAGAAACCUCCUGUCGCCAGCGGGAUUGUCAACGCCGUUGACCUCAUCUC (((((((....))))))).................(((.(((((((((.....))))..))))).)))...((((((...))))))........ (-20.09 = -21.52 + 1.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:39 2006