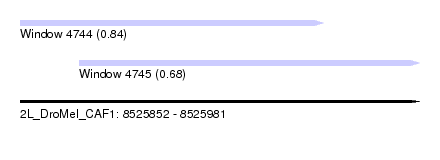
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,525,852 – 8,525,981 |
| Length | 129 |
| Max. P | 0.839313 |

| Location | 8,525,852 – 8,525,950 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 77.07 |
| Mean single sequence MFE | -19.90 |
| Consensus MFE | -12.61 |
| Energy contribution | -14.20 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.839313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8525852 98 + 22407834 AUUUUCAUCAAU-----------------GUAACUGUUACUUGGUAAAAAUUGUAUAUGUAUUUAGAAUAUAAAUAUUAUAUGCAAUGCGUUCCACAUUUACAAGGCAGUUAUCC ............-----------------((((((((..((((.((((.((((((((((((((((.....)))))).))))))))))..(.....).)))))))))))))))).. ( -21.10) >DroPse_CAF1 2574 82 + 1 -----------------------CGCU--UUCCCUUUUAUAUGGUAAAUUUUAUUUAUGUAU-------AUAAUAGUUGUAUACAGUGCGUUUCUCGACUAAAAGGCAGUUAAA- -----------------------.(((--((.....(((((((((((((...)))))).)))-------))))((((((...((.....))....)))))).))))).......- ( -12.40) >DroSec_CAF1 35050 106 + 1 GU-UUCAUCAAUU--------UUCGCUUUUUAACUGUUACUUGGUAAAAAUUGUAUAUGUAUUUAGAAUAUAAAUAUUAUAUGCAAUGCGUUCCAGACUUACAAGGCAGUUAACC ..-..........--------........((((((((...((((.....((((((((((((((((.....)))))).)))))))))).....)))).((....)))))))))).. ( -21.90) >DroSim_CAF1 34345 106 + 1 GU-UUCAUCAAUU--------UUCGCUUUUUAACUGUUACUUGGUAAAAAUUGUAUAUGUAUUUAGAAUAUAAAUAUUAUAUGCAAUGCGUUCCAGACUUACAAGGCAGUUAUCC ..-..........--------.........(((((((...((((.....((((((((((((((((.....)))))).)))))))))).....)))).((....)))))))))... ( -21.10) >DroEre_CAF1 34936 105 + 1 GUUUUUAUCAAUU--------AUCGCU--UUAACUGUUACUUGGUAAAAAUUGUAUAUGUAUUUAGGAUAUAAAUAUUAUAUGCAAUGCGUUCCAGACUUACAAGACAGUUAUCC .............--------......--.((((((((...(((.....((((((((((((((((.....)))))).)))))))))).....)))(.....)..))))))))... ( -20.90) >DroYak_CAF1 35681 113 + 1 GUUUUCAUCAAUUCGCUUUCAAUCGCU--UUAACUGUUACUUGGUAAAAAUUGUAUAUGUAUUUAGGAUAUAAAUAUUAUAUGCAAUGCGUUCCAGAGUUACAAGACAGCUAUCC (((.((........((........)).--.....(((.((((((.....((((((((((((((((.....)))))).)))))))))).....))).))).))).)).)))..... ( -22.00) >consensus GU_UUCAUCAAUU_________UCGCU__UUAACUGUUACUUGGUAAAAAUUGUAUAUGUAUUUAGAAUAUAAAUAUUAUAUGCAAUGCGUUCCAGACUUACAAGGCAGUUAUCC ..............................((((((((..((((.....((((((((((((((((.....)))))).)))))))))).....))))........))))))))... (-12.61 = -14.20 + 1.59)


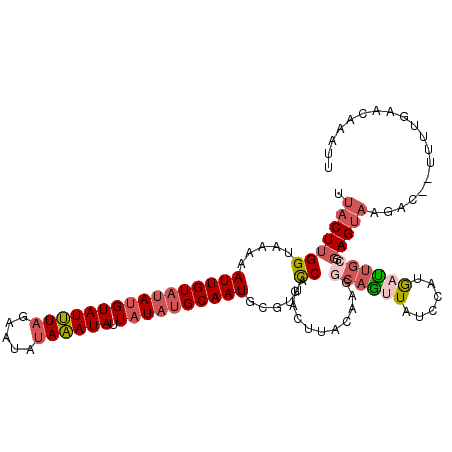
| Location | 8,525,871 – 8,525,981 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 82.05 |
| Mean single sequence MFE | -23.37 |
| Consensus MFE | -15.18 |
| Energy contribution | -16.52 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8525871 110 + 22407834 UUACUUGGUAAAAAUUGUAUAUGUAUUUAGAAUAUAAAUAUUAUAUGCAAUGCGUUCCACAUUUACAAGGCAGUUAUCCAUGAUUGCGGAGUAAGAU--UUUUGAACAAAUU ((((((((.....((((((((((((((((.....)))))).)))))))))).....))...........(((((((....))))))).))))))...--............. ( -24.60) >DroSec_CAF1 35077 110 + 1 UUACUUGGUAAAAAUUGUAUAUGUAUUUAGAAUAUAAAUAUUAUAUGCAAUGCGUUCCAGACUUACAAGGCAGUUAACCAUGAUUGCGGAGUAAGAC--UUUUGAACGAAUU .............((((((((((((((((.....)))))).)))))))))).(((((.((.(((((...(((((((....)))))))...))))).)--)...))))).... ( -27.30) >DroSim_CAF1 34372 112 + 1 UUACUUGGUAAAAAUUGUAUAUGUAUUUAGAAUAUAAAUAUUAUAUGCAAUGCGUUCCAGACUUACAAGGCAGUUAUCCAUGACUGCGGAGUAAGACAAAUUUGAACAAAUU ....((((.....((((((((((((((((.....)))))).)))))))))).....)))).(((((...(((((((....)))))))...)))))................. ( -28.60) >DroEre_CAF1 34962 109 + 1 UUACUUGGUAAAAAUUGUAUAUGUAUUUAGGAUAUAAAUAUUAUAUGCAAUGCGUUCCAGACUUACAAGACAGUUAUCC-UAAUUGCGGAGUACAAU--UUUUGAACAAAAU ....(((.((((((((((((.((((.(((((((...........((((...))))....((((........))))))))-))).))))..)))))))--)))))..)))... ( -23.50) >DroYak_CAF1 35715 110 + 1 UUACUUGGUAAAAAUUGUAUAUGUAUUUAGGAUAUAAAUAUUAUAUGCAAUGCGUUCCAGAGUUACAAGACAGCUAUCCCCAAUUGCGGAGUACGAC--UUUUGAACAAAUU ....((((.....((((((((((((((((.....)))))).)))))))))).....)))).(((.(((((..(...(((.(....).)))...)...--))))))))..... ( -22.10) >DroAna_CAF1 18649 105 + 1 UUA--UGGUAAAAAUUGUAUACGUAUAUUUAGUAUAUAUAUUUUUAGCAAUGACUUUCAGAACUGUAAAGAUAUUAUUCAAGUUUCCU---CCUGGU--UUUAGAUCCACUU ...--((.((((((((((((((.........))))))).))))))).))....((((((....)).))))..................---..(((.--.......)))... ( -14.10) >consensus UUACUUGGUAAAAAUUGUAUAUGUAUUUAGAAUAUAAAUAUUAUAUGCAAUGCGUUCCAGACUUACAAGGCAGUUAUCCAUGAUUGCGGAGUAAGAC__UUUUGAACAAAUU .(((((((.....((((((((((((((((.....)))))).)))))))))).....))...........((((((......)))))).)))))................... (-15.18 = -16.52 + 1.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:22 2006