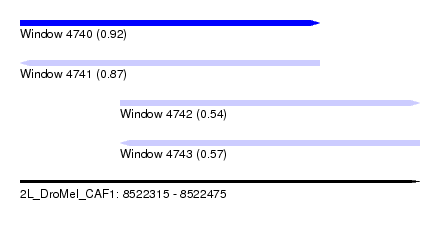
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,522,315 – 8,522,475 |
| Length | 160 |
| Max. P | 0.919132 |

| Location | 8,522,315 – 8,522,435 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.85 |
| Mean single sequence MFE | -28.70 |
| Consensus MFE | -16.59 |
| Energy contribution | -16.90 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8522315 120 + 22407834 CAUGUUGUGGGUGAGUUAUAUACGAACACAAGCCGCAAAGAUUUCGAUUAUGUUCUAUAUUUAAACAUUUGUAUAUAUUUCAUUAGCUACCCCAUGGUCUGUCCAUGGGCAAUAGCUGCU .........((((((.((((((((((((.((..((.((....)))).)).)))))((....)).......))))))).))))))(((((.(((((((.....)))))))...)))))... ( -29.00) >DroSec_CAF1 31559 120 + 1 CAUGUUGUGGGUGAGUUAUAUGCGAACACAAACCGCAAGGAAUUCGAUUAUGUUCUAUAUUUAAACACAUAUAUAUAUUUCAUUAGCUACCCCAUGGUCUGUCCAUGGGCAAUAGCUGCU .((((((((..(((((....((((.........)))).(((((........)))))..)))))..)))).)))).........((((((.(((((((.....)))))))...)))))).. ( -32.00) >DroSim_CAF1 30856 120 + 1 CAUGUUGUGGGUGAGUUAUAUACGAACACAAACCGCAAGGAAUUCGAUUAUGUUCUAUAUUUAAACACUUAUAUAUAUUUCAUUAGCUACCCCAUGGUCUAUCCAUGGGCAAUAGCUGCU .((((((((((((....(((((.(((((....((....))(((...))).)))))))))).....)))))))).)))).....((((((.(((((((.....)))))))...)))))).. ( -33.00) >DroEre_CAF1 31415 114 + 1 GAUGUUGUGGGUGAGUUGUA--CCAUCACGAACCGCGUGGAAUUCUCUUAAGUUCUAUAUUUAAACA----UGUAUAAUUUAUUAGCUACCCCAUGGACUCUCCAUGGGCAAUACCUGCU ......(..(((((((((((--(..(((((.....))))).......((((((.....))))))...----.))))))))).........(((((((.....)))))))....)))..). ( -31.20) >DroYak_CAF1 32144 118 + 1 CAUGUUGUGGGUGAGUUAUA--ACAUCACAAGCCCCAAAGAAUUCCCUAUAUUUCUAUAUUUAAACAAACGUAUAUAUUUUAUUAGCUACCCCAUGGACUGUCCAUGGGCAAUAGCUGCU .(((((((((((..((....--.....))..))))...((((..........))))........)).)))))...........((((((.(((((((.....)))))))...)))))).. ( -26.50) >DroAna_CAF1 15347 101 + 1 CUUGCUAUGGGUAUGUAUU---CC-----AG-----ACAUCCUUACAUAUAGUUCUAUAUUUAUACUUGC------GUGUUAUUAGCUGCCCCAUGGCCUGUCCAUGGGUAAUUCUUGCU ...((((((((.......)---))-----)(-----((((.(.((.(((((....))))).)).....).------)))))..))))...(((((((.....)))))))........... ( -20.50) >consensus CAUGUUGUGGGUGAGUUAUA__CCAACACAAACCGCAAAGAAUUCGAUUAAGUUCUAUAUUUAAACACAU_UAUAUAUUUCAUUAGCUACCCCAUGGUCUGUCCAUGGGCAAUAGCUGCU ....(((((.................)))))....................................................((((((.(((((((.....)))))))...)))))).. (-16.59 = -16.90 + 0.31)

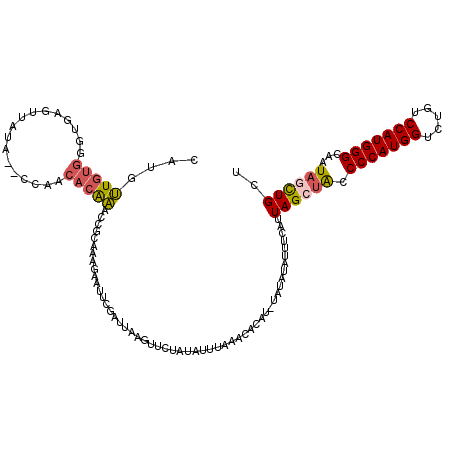
| Location | 8,522,315 – 8,522,435 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.85 |
| Mean single sequence MFE | -28.82 |
| Consensus MFE | -15.10 |
| Energy contribution | -15.93 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8522315 120 - 22407834 AGCAGCUAUUGCCCAUGGACAGACCAUGGGGUAGCUAAUGAAAUAUAUACAAAUGUUUAAAUAUAGAACAUAAUCGAAAUCUUUGCGGCUUGUGUUCGUAUAUAACUCACCCACAACAUG ...((((((..(((((((.....)))))))))))))..(((..(((((((..((((....)))).(((((((((((.........))).)))))))))))))))..)))........... ( -34.00) >DroSec_CAF1 31559 120 - 1 AGCAGCUAUUGCCCAUGGACAGACCAUGGGGUAGCUAAUGAAAUAUAUAUAUGUGUUUAAAUAUAGAACAUAAUCGAAUUCCUUGCGGUUUGUGUUCGCAUAUAACUCACCCACAACAUG ...((((((..(((((((.....)))))))))))))..(((..(((((.(((((......)))))(((((((((((.........))).))))))))..)))))..)))........... ( -31.70) >DroSim_CAF1 30856 120 - 1 AGCAGCUAUUGCCCAUGGAUAGACCAUGGGGUAGCUAAUGAAAUAUAUAUAAGUGUUUAAAUAUAGAACAUAAUCGAAUUCCUUGCGGUUUGUGUUCGUAUAUAACUCACCCACAACAUG ...((((((..(((((((.....)))))))))))))..(((..(((((((..((((....)))).(((((((((((.........))).)))))))))))))))..)))........... ( -32.00) >DroEre_CAF1 31415 114 - 1 AGCAGGUAUUGCCCAUGGAGAGUCCAUGGGGUAGCUAAUAAAUUAUACA----UGUUUAAAUAUAGAACUUAAGAGAAUUCCACGCGGUUCGUGAUGG--UACAACUCACCCACAACAUC ....(((....(((((((.....)))))))...................----....................(((....(((((((...)))).)))--.....))))))......... ( -24.20) >DroYak_CAF1 32144 118 - 1 AGCAGCUAUUGCCCAUGGACAGUCCAUGGGGUAGCUAAUAAAAUAUAUACGUUUGUUUAAAUAUAGAAAUAUAGGGAAUUCUUUGGGGCUUGUGAUGU--UAUAACUCACCCACAACAUG ...((((((..(((((((.....)))))))))))))..............(((.((((...((((....))))..))))....((((..(((((....--)))))....)))).)))... ( -28.00) >DroAna_CAF1 15347 101 - 1 AGCAAGAAUUACCCAUGGACAGGCCAUGGGGCAGCUAAUAACAC------GCAAGUAUAAAUAUAGAACUAUAUGUAAGGAUGU-----CU-----GG---AAUACAUACCCAUAGCAAG ...........(((((((.....)))))))...((((.....((------....))...............((((((.((....-----))-----..---..))))))....))))... ( -23.00) >consensus AGCAGCUAUUGCCCAUGGACAGACCAUGGGGUAGCUAAUAAAAUAUAUA_AAAUGUUUAAAUAUAGAACAUAAUCGAAUUCCUUGCGGUUUGUGUUCG__UAUAACUCACCCACAACAUG ...((((((..(((((((.....))))))))))))).................................................................................... (-15.10 = -15.93 + 0.83)

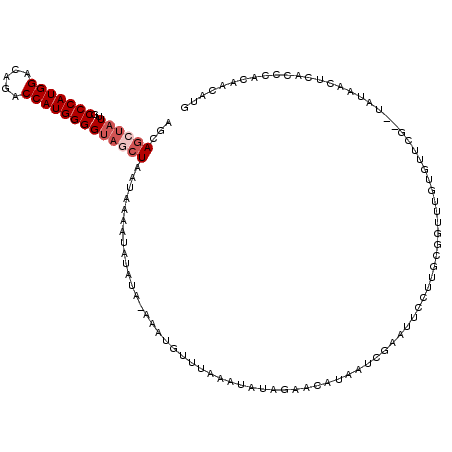

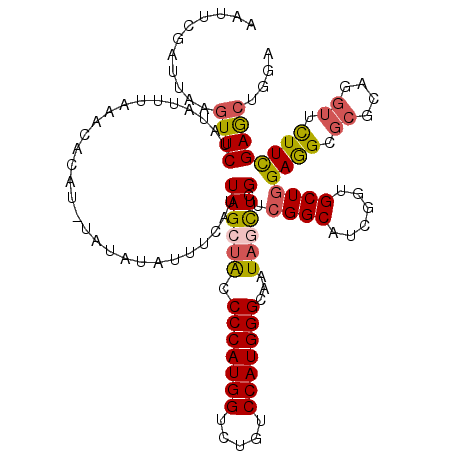
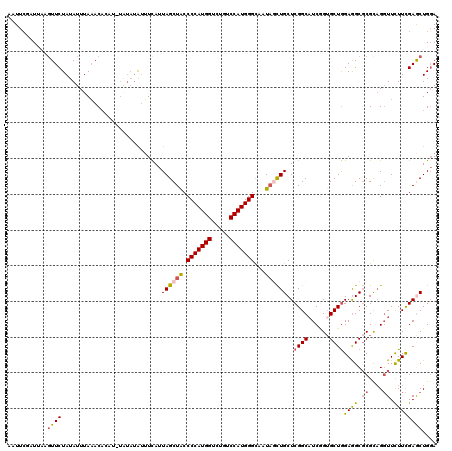
| Location | 8,522,355 – 8,522,475 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.16 |
| Mean single sequence MFE | -33.23 |
| Consensus MFE | -24.31 |
| Energy contribution | -24.48 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8522355 120 + 22407834 AUUUCGAUUAUGUUCUAUAUUUAAACAUUUGUAUAUAUUUCAUUAGCUACCCCAUGGUCUGUCCAUGGGCAAUAGCUGCUCGGCAUCGGUGCUGGAGGCGCGUAGGUUCUUCGAGCUGGA ...(((((.(((...(((((.(((....))).)))))...)))((((((.(((((((.....)))))))...))))))......))))).(((.((((.((....)).)))).))).... ( -31.70) >DroSec_CAF1 31599 120 + 1 AAUUCGAUUAUGUUCUAUAUUUAAACACAUAUAUAUAUUUCAUUAGCUACCCCAUGGUCUGUCCAUGGGCAAUAGCUGCUCGGCAUCGGUGCUGGAGGCGCGCAGGUUCUUCGAACUGGA ...........((((............................((((((.(((((((.....)))))))...))))))..(((((....)))))((((.((....)).)))))))).... ( -30.60) >DroSim_CAF1 30896 120 + 1 AAUUCGAUUAUGUUCUAUAUUUAAACACUUAUAUAUAUUUCAUUAGCUACCCCAUGGUCUAUCCAUGGGCAAUAGCUGCUCGGCAUCGGAGCUGGAAGCGCGCAGGUUCUUCGAGCUGGA ..((((((.(((...(((((.(((....))).)))))...)))((((((.(((((((.....)))))))...))))))......))))))(((.((((.((....)).)))).))).... ( -34.50) >DroEre_CAF1 31453 116 + 1 AAUUCUCUUAAGUUCUAUAUUUAAACA----UGUAUAAUUUAUUAGCUACCCCAUGGACUCUCCAUGGGCAAUACCUGCUCGGCAUCUGUGCUAGAGGCGCGCAGGUUUUUCGAGCUGCA ........((((((.(((((......)----)))).))))))...((...(((((((.....)))))))........((((((.((((((((.......))))))))...)))))).)). ( -36.30) >DroYak_CAF1 32182 120 + 1 AAUUCCCUAUAUUUCUAUAUUUAAACAAACGUAUAUAUUUUAUUAGCUACCCCAUGGACUGUCCAUGGGCAAUAGCUGCUCGGCAUCGGUGCUGGAGGCGCGCAGGUUUUUCGAGCUGGA ...(((......................................(((((.(((((((.....)))))))...)))))((((((.(((.((((.......)))).)))...)))))).))) ( -33.40) >DroAna_CAF1 15374 114 + 1 CCUUACAUAUAGUUCUAUAUUUAUACUUGC------GUGUUAUUAGCUGCCCCAUGGCCUGUCCAUGGGUAAUUCUUGCUCGGCCUCAGUGCUGGAGGCCCGCCGCUUCUUUGAACUGGA .........((((((.......((((....------))))....(((.(((((((((.....)))))))............((((((.......)))))).)).))).....)))))).. ( -32.90) >consensus AAUUCGAUUAAGUUCUAUAUUUAAACACAU_UAUAUAUUUCAUUAGCUACCCCAUGGUCUGUCCAUGGGCAAUAGCUGCUCGGCAUCGGUGCUGGAGGCGCGCAGGUUCUUCGAGCUGGA ...........((((............................((((((.(((((((.....)))))))...))))))..((((......))))((((.((....)).)))))))).... (-24.31 = -24.48 + 0.17)

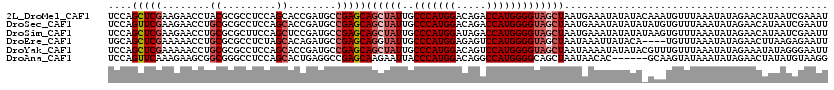
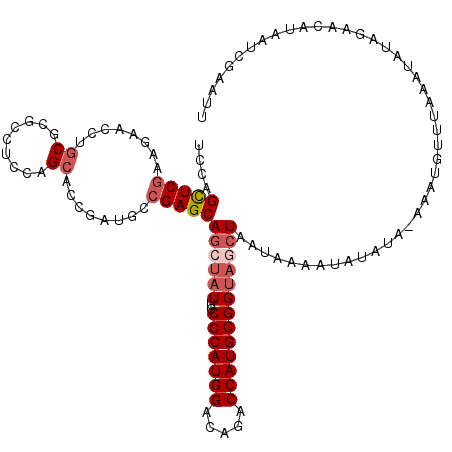
| Location | 8,522,355 – 8,522,475 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.16 |
| Mean single sequence MFE | -32.62 |
| Consensus MFE | -21.80 |
| Energy contribution | -22.74 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8522355 120 - 22407834 UCCAGCUCGAAGAACCUACGCGCCUCCAGCACCGAUGCCGAGCAGCUAUUGCCCAUGGACAGACCAUGGGGUAGCUAAUGAAAUAUAUACAAAUGUUUAAAUAUAGAACAUAAUCGAAAU ....(((((..(......)((.......))........)))))((((((..(((((((.....)))))))))))))................((((((.......))))))......... ( -29.20) >DroSec_CAF1 31599 120 - 1 UCCAGUUCGAAGAACCUGCGCGCCUCCAGCACCGAUGCCGAGCAGCUAUUGCCCAUGGACAGACCAUGGGGUAGCUAAUGAAAUAUAUAUAUGUGUUUAAAUAUAGAACAUAAUCGAAUU ...(((((((...........((.((..(((....))).))))((((((..(((((((.....)))))))))))))...............(((((((.......))))))).))))))) ( -31.70) >DroSim_CAF1 30896 120 - 1 UCCAGCUCGAAGAACCUGCGCGCUUCCAGCUCCGAUGCCGAGCAGCUAUUGCCCAUGGAUAGACCAUGGGGUAGCUAAUGAAAUAUAUAUAAGUGUUUAAAUAUAGAACAUAAUCGAAUU ...((((.((((..(....)..)))).)))).((((.......((((((..(((((((.....)))))))))))))................((((((.......)))))).)))).... ( -31.60) >DroEre_CAF1 31453 116 - 1 UGCAGCUCGAAAAACCUGCGCGCCUCUAGCACAGAUGCCGAGCAGGUAUUGCCCAUGGAGAGUCCAUGGGGUAGCUAAUAAAUUAUACA----UGUUUAAAUAUAGAACUUAAGAGAAUU ...(((((.....((((((..((.(((.....))).))...))))))....(((((((.....)))))))).)))).............----.((((.......))))........... ( -31.10) >DroYak_CAF1 32182 120 - 1 UCCAGCUCGAAAAACCUGCGCGCCUCCAGCACCGAUGCCGAGCAGCUAUUGCCCAUGGACAGUCCAUGGGGUAGCUAAUAAAAUAUAUACGUUUGUUUAAAUAUAGAAAUAUAGGGAAUU (((.(((((.......(((.........))).......)))))((((((..(((((((.....))))))))))))).........((((..(((((......)))))..)))).)))... ( -34.54) >DroAna_CAF1 15374 114 - 1 UCCAGUUCAAAGAAGCGGCGGGCCUCCAGCACUGAGGCCGAGCAAGAAUUACCCAUGGACAGGCCAUGGGGCAGCUAAUAACAC------GCAAGUAUAAAUAUAGAACUAUAUGUAAGG ...(((((.....(((.((.((((((.......))))))............(((((((.....))))))))).)))......((------....)).........))))).......... ( -37.60) >consensus UCCAGCUCGAAGAACCUGCGCGCCUCCAGCACCGAUGCCGAGCAGCUAUUGCCCAUGGACAGACCAUGGGGUAGCUAAUAAAAUAUAUA_AAAUGUUUAAAUAUAGAACAUAAUCGAAUU ....(((((........((.........))........)))))((((((..(((((((.....)))))))))))))............................................ (-21.80 = -22.74 + 0.94)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:20 2006