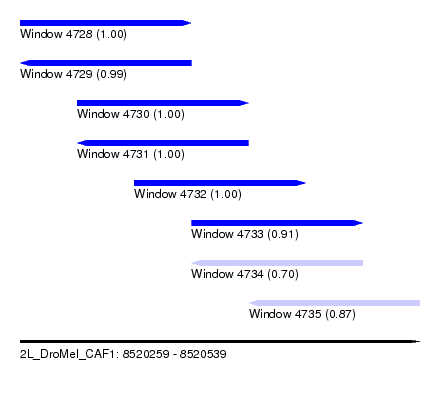
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,520,259 – 8,520,539 |
| Length | 280 |
| Max. P | 0.999943 |

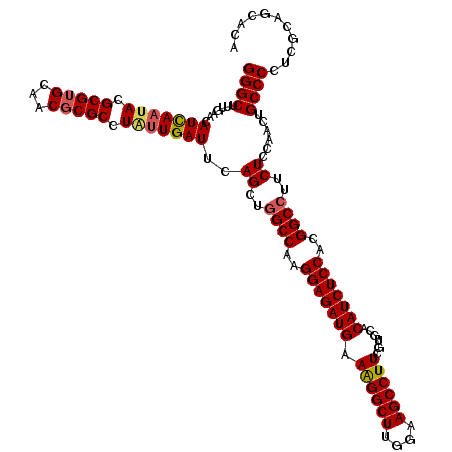
| Location | 8,520,259 – 8,520,379 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.75 |
| Mean single sequence MFE | -42.88 |
| Consensus MFE | -39.56 |
| Energy contribution | -39.76 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.36 |
| SVM RNA-class probability | 0.999881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8520259 120 + 22407834 GGGCUUUGAACAUUAAUACGCGUGCAACGCGCCUGAUGAUUCAGCUGGCCAAGGAGAUGAAAGGCUUGGAAGCCUUCGUGCACAUCUCCACGGCCUUCUCCAACUGCCCCUCGCAGCACA ((((.((((((((((...((((.....))))..))))).)))))..((((..(((((((.((((((....))))))......)))))))..))))..........))))........... ( -43.10) >DroSec_CAF1 29503 120 + 1 GGGCUUUGAAUAUCAAUACGCGUGCAACGCGCCUUUUGAUUCAGCUGGCCAAGGAGAUGAAAGGCUUGAAAGCCUUCGUCCACAUCUCCACGGCCUUCUCCAACUGCCCCUCGCAGCACA ((((.((((..(((((...(((((...)))))...)))))))))..((((..(((((((.((((((....))))))......)))))))..))))..........))))........... ( -41.50) >DroSim_CAF1 28837 120 + 1 GGGCUUUGAAUAUCAAUACGCGUGCAACGCGCCUAUUGAUUCAGCUUGCCAAGGAGAUGAAAGGCUUGAAAGCCUUCGUCCACAUCUCCACGGCCUUCUCCAACUGCCCCUCGCAGCAUA ((((.((((..(((((((.(((((...))))).)))))))))))...(((..(((((((.((((((....))))))......)))))))..)))...........))))........... ( -41.10) >DroEre_CAF1 29121 120 + 1 GGGCUGUUAACAUUAAUACGCGGGCCACGCGCCUAUUGAUUCAGCUGGCCAAGGAGAUGAAGGGCUUAGAAGCCUUCGUGCACAUCUCCACGGCCUUCUCCAACUGCCCCUCGCAGCACA ((((((.....(((((((((((.....))))..))))))).)))).((((..(((((((.((((((....))))))......)))))))..))))....))..((((.....)))).... ( -42.60) >DroYak_CAF1 30000 120 + 1 GGGCGCUCAACAUCAAUACGCGUGGCACGCGCCUAUUGAUUCAGCUGGCCAAGGAGAUGAAGGGCUUAGAAGCCUUCGUGCACAUCUCCACGGCCUUCUCCAACUGCCCCUCGCAGCACA (((((((....(((((((.(((((...))))).)))))))..))).((((..(((((((.((((((....))))))......)))))))..))))..........))))........... ( -46.10) >consensus GGGCUUUGAACAUCAAUACGCGUGCAACGCGCCUAUUGAUUCAGCUGGCCAAGGAGAUGAAAGGCUUGGAAGCCUUCGUGCACAUCUCCACGGCCUUCUCCAACUGCCCCUCGCAGCACA ((((.......(((((((.(((((...))))).)))))))..((..((((..(((((((.((((((....))))))......)))))))..))))..))......))))........... (-39.56 = -39.76 + 0.20)

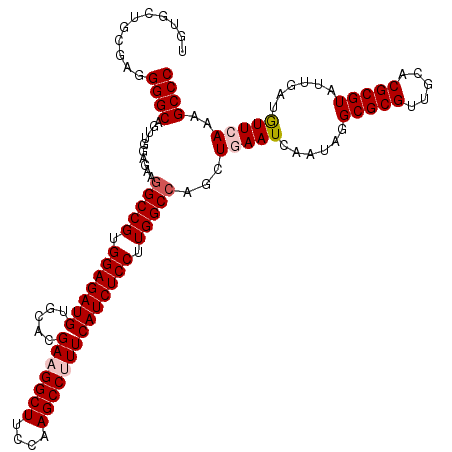
| Location | 8,520,259 – 8,520,379 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.75 |
| Mean single sequence MFE | -46.78 |
| Consensus MFE | -42.16 |
| Energy contribution | -42.92 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8520259 120 - 22407834 UGUGCUGCGAGGGGCAGUUGGAGAAGGCCGUGGAGAUGUGCACGAAGGCUUCCAAGCCUUUCAUCUCCUUGGCCAGCUGAAUCAUCAGGCGCGUUGCACGCGUAUUAAUGUUCAAAGCCC ...........((((..(((((.(.(((((.(((((((.....(((((((....)))))))))))))).)))))..((((....))))(((((.....))))).....).))))).)))) ( -48.70) >DroSec_CAF1 29503 120 - 1 UGUGCUGCGAGGGGCAGUUGGAGAAGGCCGUGGAGAUGUGGACGAAGGCUUUCAAGCCUUUCAUCUCCUUGGCCAGCUGAAUCAAAAGGCGCGUUGCACGCGUAUUGAUAUUCAAAGCCC ...........((((..(((((...(((((.(((((((.....(((((((....)))))))))))))).)))))......(((((...(((((.....))))).))))).))))).)))) ( -49.30) >DroSim_CAF1 28837 120 - 1 UAUGCUGCGAGGGGCAGUUGGAGAAGGCCGUGGAGAUGUGGACGAAGGCUUUCAAGCCUUUCAUCUCCUUGGCAAGCUGAAUCAAUAGGCGCGUUGCACGCGUAUUGAUAUUCAAAGCCC ...........((((..(((((....((((.(((((((.....(((((((....)))))))))))))).)))).......(((((((.(((.......))).))))))).))))).)))) ( -46.20) >DroEre_CAF1 29121 120 - 1 UGUGCUGCGAGGGGCAGUUGGAGAAGGCCGUGGAGAUGUGCACGAAGGCUUCUAAGCCCUUCAUCUCCUUGGCCAGCUGAAUCAAUAGGCGCGUGGCCCGCGUAUUAAUGUUAACAGCCC ...(((((.....))))).((....(((((.(((((((.....((.((((....)))).))))))))).))))).((((...((.((((((((.....))))).))).))....)))))) ( -42.50) >DroYak_CAF1 30000 120 - 1 UGUGCUGCGAGGGGCAGUUGGAGAAGGCCGUGGAGAUGUGCACGAAGGCUUCUAAGCCCUUCAUCUCCUUGGCCAGCUGAAUCAAUAGGCGCGUGCCACGCGUAUUGAUGUUGAGCGCCC ...........((((..........(((((.(((((((.....((.((((....)))).))))))))).))))).(((..(((((((.(((.......))).)))))))....))))))) ( -47.20) >consensus UGUGCUGCGAGGGGCAGUUGGAGAAGGCCGUGGAGAUGUGCACGAAGGCUUCCAAGCCUUUCAUCUCCUUGGCCAGCUGAAUCAAUAGGCGCGUUGCACGCGUAUUGAUGUUCAAAGCCC ...........((((..........(((((.(((((((.....(((((((....)))))))))))))).)))))...(((((......(((((.....)))))......)))))..)))) (-42.16 = -42.92 + 0.76)

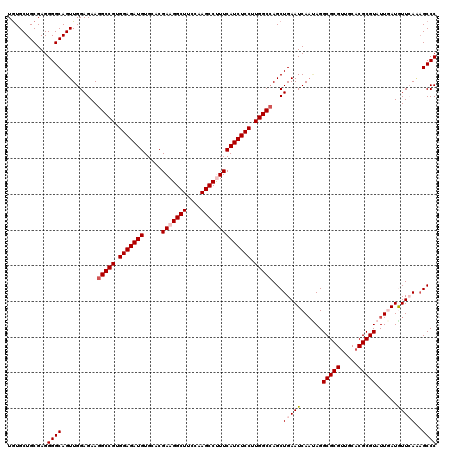
| Location | 8,520,299 – 8,520,419 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -44.00 |
| Consensus MFE | -42.78 |
| Energy contribution | -42.94 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.75 |
| SVM RNA-class probability | 0.999583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8520299 120 + 22407834 UCAGCUGGCCAAGGAGAUGAAAGGCUUGGAAGCCUUCGUGCACAUCUCCACGGCCUUCUCCAACUGCCCCUCGCAGCACAUUGAGGAGCGUUUCUAUCCGGAGCACCUAACGUGCCCGGC ..((..((((..(((((((.((((((....))))))......)))))))..))))..))......((.(((((........))))).))........((((.((((.....)))))))). ( -45.20) >DroSec_CAF1 29543 120 + 1 UCAGCUGGCCAAGGAGAUGAAAGGCUUGAAAGCCUUCGUCCACAUCUCCACGGCCUUCUCCAACUGCCCCUCGCAGCACAUUGAGGAGCGUUUCUAUCCGGAGCACCUAUCGUGCCCGAC ..((..((((..(((((((.((((((....))))))......)))))))..))))..))......((.(((((........))))).)).........(((.((((.....))))))).. ( -42.80) >DroSim_CAF1 28877 120 + 1 UCAGCUUGCCAAGGAGAUGAAAGGCUUGAAAGCCUUCGUCCACAUCUCCACGGCCUUCUCCAACUGCCCCUCGCAGCAUAUUGAGGAGCGUUUCUAUCCGGAGCACCUAUCGUGCCCGGC ...(((.(((..(((((((.((((((....))))))......)))))))..)))((((.....((((.....))))......)))))))........((((.((((.....)))))))). ( -42.20) >DroEre_CAF1 29161 120 + 1 UCAGCUGGCCAAGGAGAUGAAGGGCUUAGAAGCCUUCGUGCACAUCUCCACGGCCUUCUCCAACUGCCCCUCGCAGCACAUUGAGGAGCGUUUCUAUCCGGAGCACCUGUCGUGCCCGGC ..((..((((..(((((((.((((((....))))))......)))))))..))))..))......((.(((((........))))).))........((((.((((.....)))))))). ( -44.90) >DroYak_CAF1 30040 120 + 1 UCAGCUGGCCAAGGAGAUGAAGGGCUUAGAAGCCUUCGUGCACAUCUCCACGGCCUUCUCCAACUGCCCCUCGCAGCACAUUGAGGAGCGUUUCUAUCCGGAGCACCUAUCGUGCCCGGC ..((..((((..(((((((.((((((....))))))......)))))))..))))..))......((.(((((........))))).))........((((.((((.....)))))))). ( -44.90) >consensus UCAGCUGGCCAAGGAGAUGAAAGGCUUGGAAGCCUUCGUGCACAUCUCCACGGCCUUCUCCAACUGCCCCUCGCAGCACAUUGAGGAGCGUUUCUAUCCGGAGCACCUAUCGUGCCCGGC ..((..((((..(((((((.((((((....))))))......)))))))..))))..))......((.(((((........))))).))........((((.((((.....)))))))). (-42.78 = -42.94 + 0.16)

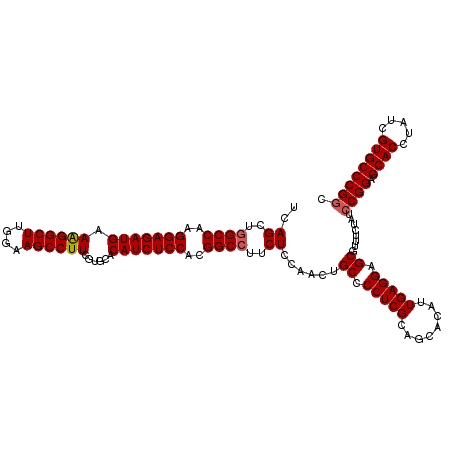
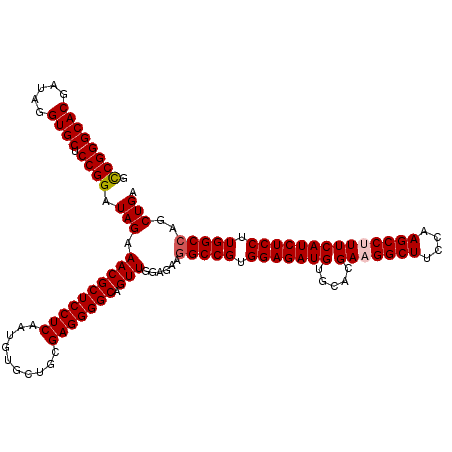
| Location | 8,520,299 – 8,520,419 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -52.10 |
| Consensus MFE | -51.50 |
| Energy contribution | -51.94 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.02 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.72 |
| SVM RNA-class probability | 0.999943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8520299 120 - 22407834 GCCGGGCACGUUAGGUGCUCCGGAUAGAAACGCUCCUCAAUGUGCUGCGAGGGGCAGUUGGAGAAGGCCGUGGAGAUGUGCACGAAGGCUUCCAAGCCUUUCAUCUCCUUGGCCAGCUGA .((((((((.....)))).)))).....((((((((((..........))))))).)))..((..(((((.(((((((.....(((((((....)))))))))))))).)))))..)).. ( -54.80) >DroSec_CAF1 29543 120 - 1 GUCGGGCACGAUAGGUGCUCCGGAUAGAAACGCUCCUCAAUGUGCUGCGAGGGGCAGUUGGAGAAGGCCGUGGAGAUGUGGACGAAGGCUUUCAAGCCUUUCAUCUCCUUGGCCAGCUGA .((((((((.....)))).)))).....((((((((((..........))))))).)))..((..(((((.(((((((.....(((((((....)))))))))))))).)))))..)).. ( -52.20) >DroSim_CAF1 28877 120 - 1 GCCGGGCACGAUAGGUGCUCCGGAUAGAAACGCUCCUCAAUAUGCUGCGAGGGGCAGUUGGAGAAGGCCGUGGAGAUGUGGACGAAGGCUUUCAAGCCUUUCAUCUCCUUGGCAAGCUGA .((((((((.....)))).)))).....((((((((((..........))))))).)))..((...((((.(((((((.....(((((((....)))))))))))))).))))...)).. ( -52.10) >DroEre_CAF1 29161 120 - 1 GCCGGGCACGACAGGUGCUCCGGAUAGAAACGCUCCUCAAUGUGCUGCGAGGGGCAGUUGGAGAAGGCCGUGGAGAUGUGCACGAAGGCUUCUAAGCCCUUCAUCUCCUUGGCCAGCUGA .((((((((.....)))).)))).....((((((((((..........))))))).)))..((..(((((.(((((((.....((.((((....)))).))))))))).)))))..)).. ( -50.70) >DroYak_CAF1 30040 120 - 1 GCCGGGCACGAUAGGUGCUCCGGAUAGAAACGCUCCUCAAUGUGCUGCGAGGGGCAGUUGGAGAAGGCCGUGGAGAUGUGCACGAAGGCUUCUAAGCCCUUCAUCUCCUUGGCCAGCUGA .((((((((.....)))).)))).....((((((((((..........))))))).)))..((..(((((.(((((((.....((.((((....)))).))))))))).)))))..)).. ( -50.70) >consensus GCCGGGCACGAUAGGUGCUCCGGAUAGAAACGCUCCUCAAUGUGCUGCGAGGGGCAGUUGGAGAAGGCCGUGGAGAUGUGCACGAAGGCUUCCAAGCCUUUCAUCUCCUUGGCCAGCUGA .((((((((.....)))).)))).(((.((((((((((..........))))))).)))......(((((.(((((((.....(((((((....)))))))))))))).)))))..))). (-51.50 = -51.94 + 0.44)

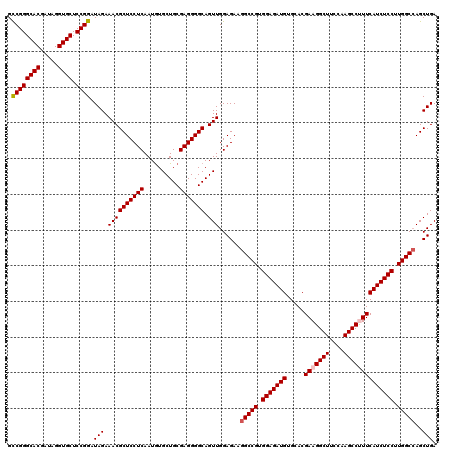
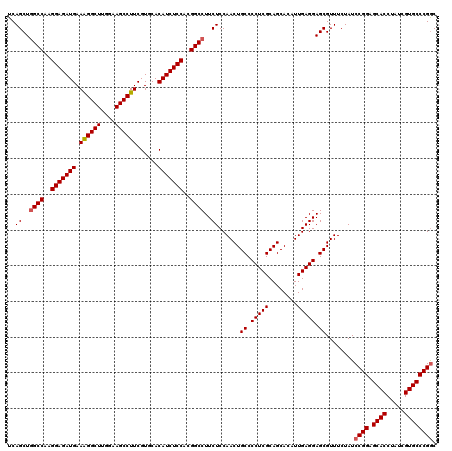
| Location | 8,520,339 – 8,520,459 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -41.98 |
| Consensus MFE | -38.96 |
| Energy contribution | -39.76 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.52 |
| SVM RNA-class probability | 0.999328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8520339 120 + 22407834 CACAUCUCCACGGCCUUCUCCAACUGCCCCUCGCAGCACAUUGAGGAGCGUUUCUAUCCGGAGCACCUAACGUGCCCGGCGGCCAAGGUGCUCGAAUUCAACGAGACUUUGAGCCCAGAU ...((((....((((..........((.(((((........))))).))........((((.((((.....)))))))).))))..((.(((((((.((.....)).))))))))))))) ( -43.00) >DroSec_CAF1 29583 120 + 1 CACAUCUCCACGGCCUUCUCCAACUGCCCCUCGCAGCACAUUGAGGAGCGUUUCUAUCCGGAGCACCUAUCGUGCCCGACGGCCAAGGUGCUCGAAUUCAACGAGACUUUGAGCCCAGAU ...((((....((((..........((.(((((........))))).)).........(((.((((.....)))))))..))))..((.(((((((.((.....)).))))))))))))) ( -40.60) >DroSim_CAF1 28917 120 + 1 CACAUCUCCACGGCCUUCUCCAACUGCCCCUCGCAGCAUAUUGAGGAGCGUUUCUAUCCGGAGCACCUAUCGUGCCCGGCGGCCAAGGUGCUCGAAUUCAACGAGACUUUGAGCCCAGAU ...((((....((((..........((.(((((........))))).))........((((.((((.....)))))))).))))..((.(((((((.((.....)).))))))))))))) ( -43.00) >DroEre_CAF1 29201 120 + 1 CACAUCUCCACGGCCUUCUCCAACUGCCCCUCGCAGCACAUUGAGGAGCGUUUCUAUCCGGAGCACCUGUCGUGCCCGGCGGCCAAGGUGCUCGAACUUAACGACACUUUGAGCCCAGAC ....(((....((((..........((.(((((........))))).))........((((.((((.....)))))))).))))..((.(((((((...........)))))))))))). ( -39.40) >DroYak_CAF1 30080 120 + 1 CACAUCUCCACGGCCUUCUCCAACUGCCCCUCGCAGCACAUUGAGGAGCGUUUCUAUCCGGAGCACCUAUCGUGCCCGGCGGCCAAGGUGCUCGAAUUCAACGAGACUUUGAGCGCAGAU ...((((....((((..........((.(((((........))))).))........((((.((((.....)))))))).))))...(((((((((.((.....)).))))))))))))) ( -43.90) >consensus CACAUCUCCACGGCCUUCUCCAACUGCCCCUCGCAGCACAUUGAGGAGCGUUUCUAUCCGGAGCACCUAUCGUGCCCGGCGGCCAAGGUGCUCGAAUUCAACGAGACUUUGAGCCCAGAU ...((((....((((..........((.(((((........))))).))........((((.((((.....)))))))).))))..((.(((((((.((.....)).))))))))))))) (-38.96 = -39.76 + 0.80)

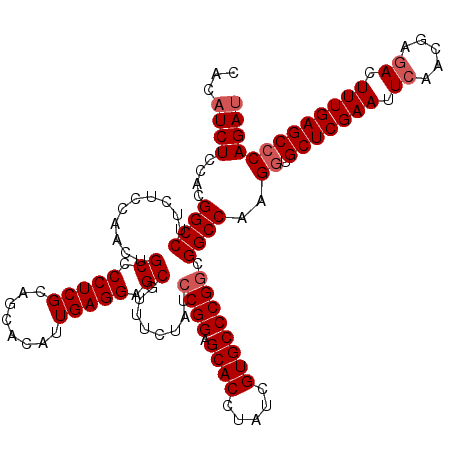
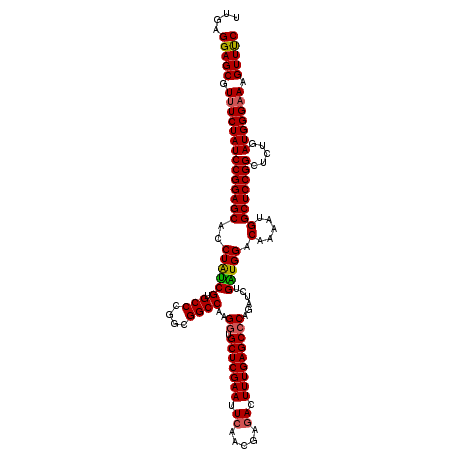
| Location | 8,520,379 – 8,520,499 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -44.69 |
| Consensus MFE | -39.58 |
| Energy contribution | -39.30 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.912921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8520379 120 + 22407834 UUGAGGAGCGUUUCUAUCCGGAGCACCUAACGUGCCCGGCGGCCAAGGUGCUCGAAUUCAACGAGACUUUGAGCCCAGAUUUGUUGGACAAGAUGGCUCCGGCUCUUAUGGGAAAGUUUC ....(((((.((((((((((((((........((.((((((((((((((.((((.......)))))))))).))).......))))).)).....))))))).....))))))).))))) ( -42.13) >DroSec_CAF1 29623 120 + 1 UUGAGGAGCGUUUCUAUCCGGAGCACCUAUCGUGCCCGACGGCCAAGGUGCUCGAAUUCAACGAGACUUUGAGCCCAGAUCUGAUGGACAAAAUGGCUCCGGCUCUGAUGGGAAAGUUUC ....(((((.(((((((((((.((((.....)))))))..((((..((.(((((((.((.....)).))))))))).........((((......).)))))))..)))))))).))))) ( -41.80) >DroSim_CAF1 28957 120 + 1 UUGAGGAGCGUUUCUAUCCGGAGCACCUAUCGUGCCCGGCGGCCAAGGUGCUCGAAUUCAACGAGACUUUGAGCCCAGAUCUGAUGGACAAAAUGGCUCCGGCUCUGAUGGGAAAGUUUC ....(((((.((((((((.(((((((.....))))((((.(((((.((.(((((((.((.....)).)))))))))...((.....)).....))))))))).))))))))))).))))) ( -43.20) >DroEre_CAF1 29241 120 + 1 UUGAGGAGCGUUUCUAUCCGGAGCACCUGUCGUGCCCGGCGGCCAAGGUGCUCGAACUUAACGACACUUUGAGCCCAGACCUGGUGGACAACAUGGCUCCGGCUCUGAUGGGAAAGUUCC ....(((((.((((((((((((((...((((.((((.(((((((((((((.(((.......)))))))))).)))..).)).)))))))).....))))))).....))))))).))))) ( -50.10) >DroYak_CAF1 30120 120 + 1 UUGAGGAGCGUUUCUAUCCGGAGCACCUAUCGUGCCCGGCGGCCAAGGUGCUCGAAUUCAACGAGACUUUGAGCGCAGAUCUGGUGGACAACAUGGCUCCGGCUCUAAUGGGCAAGUUCC ....(((((........((((.((((.....))))))))(.((((..(((((((((.((.....)).))))))))).....)))).)........))))).((((....))))....... ( -46.20) >consensus UUGAGGAGCGUUUCUAUCCGGAGCACCUAUCGUGCCCGGCGGCCAAGGUGCUCGAAUUCAACGAGACUUUGAGCCCAGAUCUGAUGGACAAAAUGGCUCCGGCUCUGAUGGGAAAGUUUC ....(((((.((((((((((((((..((((((.(((....))))..((.(((((((.((.....)).)))))))))......))))).(.....)))))))).....))))))).))))) (-39.58 = -39.30 + -0.28)

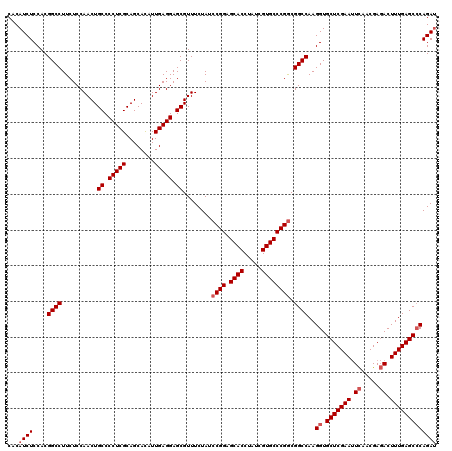
| Location | 8,520,379 – 8,520,499 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -40.76 |
| Consensus MFE | -35.62 |
| Energy contribution | -35.50 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8520379 120 - 22407834 GAAACUUUCCCAUAAGAGCCGGAGCCAUCUUGUCCAACAAAUCUGGGCUCAAAGUCUCGUUGAAUUCGAGCACCUUGGCCGCCGGGCACGUUAGGUGCUCCGGAUAGAAACGCUCCUCAA ....................(((((..(((.((((..(((....((((.....))))..))).....((((((((..(((....))).....)))))))).)))))))...))))).... ( -38.20) >DroSec_CAF1 29623 120 - 1 GAAACUUUCCCAUCAGAGCCGGAGCCAUUUUGUCCAUCAGAUCUGGGCUCAAAGUCUCGUUGAAUUCGAGCACCUUGGCCGUCGGGCACGAUAGGUGCUCCGGAUAGAAACGCUCCUCAA ....................(((((..((((((((.((((....((((.....))))..))))....((((((((..(((....))).....)))))))).))))))))..))))).... ( -41.60) >DroSim_CAF1 28957 120 - 1 GAAACUUUCCCAUCAGAGCCGGAGCCAUUUUGUCCAUCAGAUCUGGGCUCAAAGUCUCGUUGAAUUCGAGCACCUUGGCCGCCGGGCACGAUAGGUGCUCCGGAUAGAAACGCUCCUCAA ....................(((((..((((((((.((((....((((.....))))..))))....((((((((..(((....))).....)))))))).))))))))..))))).... ( -41.60) >DroEre_CAF1 29241 120 - 1 GGAACUUUCCCAUCAGAGCCGGAGCCAUGUUGUCCACCAGGUCUGGGCUCAAAGUGUCGUUAAGUUCGAGCACCUUGGCCGCCGGGCACGACAGGUGCUCCGGAUAGAAACGCUCCUCAA (((.(((((.........((((((((.((((((((....((((.((((((.((.(.......).)).)))).))..))))....)).)))))).).)))))))...)))).).))).... ( -42.50) >DroYak_CAF1 30120 120 - 1 GGAACUUGCCCAUUAGAGCCGGAGCCAUGUUGUCCACCAGAUCUGCGCUCAAAGUCUCGUUGAAUUCGAGCACCUUGGCCGCCGGGCACGAUAGGUGCUCCGGAUAGAAACGCUCCUCAA ((..((........))..))(((((....((((((.........(((..(((.((((((.......)))).)).)))..))).((((((.....)))).))))))))....))))).... ( -39.90) >consensus GAAACUUUCCCAUCAGAGCCGGAGCCAUGUUGUCCACCAGAUCUGGGCUCAAAGUCUCGUUGAAUUCGAGCACCUUGGCCGCCGGGCACGAUAGGUGCUCCGGAUAGAAACGCUCCUCAA ...............((((..(((((...(((.....))).....)))))........(((..((((((((((((..(((....))).....))))))).)))))...)))))))..... (-35.62 = -35.50 + -0.12)

| Location | 8,520,419 – 8,520,539 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.75 |
| Mean single sequence MFE | -41.44 |
| Consensus MFE | -35.88 |
| Energy contribution | -36.56 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8520419 120 - 22407834 ACCUGCUCGGCCAGUGCCUUGGUAUAGGUAUAGGUGUUCGGAAACUUUCCCAUAAGAGCCGGAGCCAUCUUGUCCAACAAAUCUGGGCUCAAAGUCUCGUUGAAUUCGAGCACCUUGGCC ........(((((((((((......))))))(((((((((((.....)))((...(((.(.(((((...(((.....))).....)))))...).)))..)).....))))))))))))) ( -41.90) >DroSec_CAF1 29663 120 - 1 ACCUGCUCGGCCAGUGCCUUGGUAUAGGUAUAGGUGUUCGGAAACUUUCCCAUCAGAGCCGGAGCCAUUUUGUCCAUCAGAUCUGGGCUCAAAGUCUCGUUGAAUUCGAGCACCUUGGCC ........(((((((((((......))))))(((((((((((.....)))((...(((.(.(((((..((((.....))))....)))))...).)))..)).....))))))))))))) ( -43.50) >DroSim_CAF1 28997 120 - 1 ACCUGCUCGGCCAGUGCCUUGGUAUAGGUAUAGGUGUUCGGAAACUUUCCCAUCAGAGCCGGAGCCAUUUUGUCCAUCAGAUCUGGGCUCAAAGUCUCGUUGAAUUCGAGCACCUUGGCC ........(((((((((((......))))))(((((((((((.....)))((...(((.(.(((((..((((.....))))....)))))...).)))..)).....))))))))))))) ( -43.50) >DroEre_CAF1 29281 120 - 1 ACCUGCUCGGCCAAUGCCUUGGUAUAUGUAUAGGUGUUCGGGAACUUUCCCAUCAGAGCCGGAGCCAUGUUGUCCACCAGGUCUGGGCUCAAAGUGUCGUUAAGUUCGAGCACCUUGGCC ........(((((((((....))))......((((((((((((....))))....(((((.(..((.((........))))..).))))).................))))))))))))) ( -43.20) >DroYak_CAF1 30160 120 - 1 ACCUGCUCAGCCAGUGCCUUGGUGUAUGUAUAGGUGUUCGGGAACUUGCCCAUUAGAGCCGGAGCCAUGUUGUCCACCAGAUCUGCGCUCAAAGUCUCGUUGAAUUCGAGCACCUUGGCC .........(((((..(....)..)......(((((((((((......)))....(((.(.((((((.(((........))).)).))))...).))).........)))))))))))). ( -35.10) >consensus ACCUGCUCGGCCAGUGCCUUGGUAUAGGUAUAGGUGUUCGGAAACUUUCCCAUCAGAGCCGGAGCCAUGUUGUCCACCAGAUCUGGGCUCAAAGUCUCGUUGAAUUCGAGCACCUUGGCC ........(((((((((((......))))))((((((((((........))....(((.(.(((((...(((.....))).....)))))...).))).........))))))))))))) (-35.88 = -36.56 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:12 2006