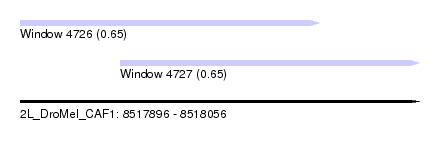
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,517,896 – 8,518,056 |
| Length | 160 |
| Max. P | 0.653925 |

| Location | 8,517,896 – 8,518,016 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.78 |
| Mean single sequence MFE | -46.72 |
| Consensus MFE | -28.97 |
| Energy contribution | -29.52 |
| Covariance contribution | 0.55 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8517896 120 + 22407834 ACUGCCCGACUUCGGCGUCCAGGGACAAUUAAAGCUGAAAAACAGCUCUGUCCUGAUUGUGGGCCUAGGCGGAUUGGGAUGUCCGGCGGCGCAAUACUUGGCGGCAGCCGGAUGCGGACA .((((((((.((((.((((((((((((.....(((((.....))))).)))))).....)))))...).))))))))...((((((((.(((........))).).)))))))))))... ( -48.90) >DroSec_CAF1 27220 120 + 1 ACUGCCCGACUUCGGCGUCCAGGGACAACUAAAGCUGAAAAACAGCUCUGUCCUGAUUGUGGGAAUGGGCGGAUUGGGAUGUCCCGCGGCGCAAUACUUGGCGGCAGCCGGAUGCGGACA .....(((..((((((.....((((((.((((.((((.....))))(((((((.............)))))))))))..))))))((.((.((.....)))).)).))))))..)))... ( -47.92) >DroSim_CAF1 26907 120 + 1 ACUGCCCGACUUCGGCGUCCAGGGACAACUAAAGCUGAAAAACAGCUCUGUCCUGAUUGUGGGAAUGGGCGGAUUGGGAUGUCCCGCGGNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN .(((((((..(((.(((....((((((.....(((((.....))))).))))))...))).))).)))))))...(((....)))................................... ( -34.90) >DroEre_CAF1 26995 120 + 1 ACUGCCCGACUUCGGGGUCCAGGGACAACUAAAGCUGAAAAACAGUUCUGUCCUGAUUGUGGGCCUGGGCGGAUUGGGAUGUCCGGCGGCGCAAUACCUGGCGGCAGCCGGAUGCGGCCG .(((((((.(....)((((((((((((.....(((((.....))))).)))))).....)))))))))))))....((.(((((((((.(((........))).).))))))))...)). ( -52.60) >DroYak_CAF1 27849 120 + 1 ACUGCCCGACUUCGGAGUCCAGGGACAACUAAAGCUGAAGAACAGCUCUGUGUUGAUUGUGGGCCUGGGCGGAUUGGGAUGUCCGGCGGCGCAAUACUUGGCGGCAGCCGGAUGUGGACG .(((((((.(....).(((((.((.((((((.(((((.....))))).)).)))).)).))))).)))))))......((((((((((.(((........))).).)))))))))..... ( -51.50) >DroAna_CAF1 13212 120 + 1 UCUCCCAGAUUUCGGUGUCCAAGGCCAGUUGAGGCUAAAGAACAGUUCCGUUCUUAUCGUUGGCAUGGGAGGAUUGGGUUGUCCGGCAGCGCAGUACCUUGCUGCAGCUGGCUGCGGAAA ...(((((.((((.(((.((((((((......)))).((((((......))))))....))))))).))))..)))))...((((.(((((((((.....))))).....)))))))).. ( -44.50) >consensus ACUGCCCGACUUCGGCGUCCAGGGACAACUAAAGCUGAAAAACAGCUCUGUCCUGAUUGUGGGCAUGGGCGGAUUGGGAUGUCCGGCGGCGCAAUACUUGGCGGCAGCCGGAUGCGGACA .(((((.(((.((...(((....)))..((((.((((.....))))(((((((.............))))))))))))).))).)))))((((....(((((....))))).)))).... (-28.97 = -29.52 + 0.55)

| Location | 8,517,936 – 8,518,056 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.58 |
| Mean single sequence MFE | -51.52 |
| Consensus MFE | -37.54 |
| Energy contribution | -36.90 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.646657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8517936 120 + 22407834 AACAGCUCUGUCCUGAUUGUGGGCCUAGGCGGAUUGGGAUGUCCGGCGGCGCAAUACUUGGCGGCAGCCGGAUGCGGACACUUGGGGCUUGUCGACUACGACGAGGUGGAGCGGAGCAAC ....(((((((((..(.(((...(((((.....)))))..((((((((.(((........))).).)))))))....))).)..)).(((((((....))))))).....)))))))... ( -49.20) >DroSec_CAF1 27260 120 + 1 AACAGCUCUGUCCUGAUUGUGGGAAUGGGCGGAUUGGGAUGUCCCGCGGCGCAAUACUUGGCGGCAGCCGGAUGCGGACACUUGGGGCUUGUCGACUACGACGAGGUGGAGCGGAGCAAC ....(((((((((..(.(((..(..(.(((.....(((....)))((.((.((.....)))).)).))).)...)..))).)..)).(((((((....))))))).....)))))))... ( -44.00) >DroEre_CAF1 27035 120 + 1 AACAGUUCUGUCCUGAUUGUGGGCCUGGGCGGAUUGGGAUGUCCGGCGGCGCAAUACCUGGCGGCAGCCGGAUGCGGCCGCCUGGGACUCAUAGACUACGACGAGGUGGAGCGGAGCAAC ....(((((((.....(((((((((..(((((.(((...(((((((((.(((........))).).))))))))))))))))..)).))))))).((((......)))).)))))))... ( -55.70) >DroYak_CAF1 27889 120 + 1 AACAGCUCUGUGUUGAUUGUGGGCCUGGGCGGAUUGGGAUGUCCGGCGGCGCAAUACUUGGCGGCAGCCGGAUGUGGACGCUUGGGACUCAUAGACUACGACGAGGUGGAGCGGAGCAAC ....(((((((.....(((((((((..((((..(....((((((((((.(((........))).).))))))))))..))))..)).))))))).((((......)))).)))))))... ( -54.50) >DroAna_CAF1 13252 120 + 1 AACAGUUCCGUUCUUAUCGUUGGCAUGGGAGGAUUGGGUUGUCCGGCAGCGCAGUACCUUGCUGCAGCUGGCUGCGGAAAGCUGGGUCUAAUCGACUACGACGAGGUGGAGCGGAGCAAU ....((((((((((..((((((...(((..(((((.((((.((((.(((((((((.....))))).....)))))))).)))).)))))..)))....))))))...))))))))))... ( -54.20) >consensus AACAGCUCUGUCCUGAUUGUGGGCCUGGGCGGAUUGGGAUGUCCGGCGGCGCAAUACUUGGCGGCAGCCGGAUGCGGACACUUGGGACUCAUCGACUACGACGAGGUGGAGCGGAGCAAC ....((((((((((..((((((...(((((...(..((.((((((.((((((...........)).))))....))))))))..).)))))....))))))..)))....)))))))... (-37.54 = -36.90 + -0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:04 2006