
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,507,185 – 8,507,280 |
| Length | 95 |
| Max. P | 0.867451 |

| Location | 8,507,185 – 8,507,280 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.97 |
| Mean single sequence MFE | -26.27 |
| Consensus MFE | -16.01 |
| Energy contribution | -16.87 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
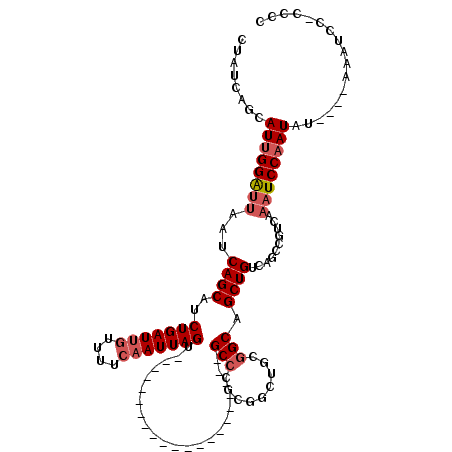
>2L_DroMel_CAF1 8507185 95 + 22407834 CUAUCAGCAUUGGAUUAAUCAGCAUCUGAUAGUUUUCAAUUAGU---------------------GCCCGCGGAUGCGGCAGCUGUCAGCCAUCAAAUCCAAUAU----AAAUCCCCCCC ........((((((((.....(((((((...((...........---------------------))...)))))))(((........)))....))))))))..----........... ( -23.00) >DroSec_CAF1 16514 98 + 1 CUAUCAGCAUUGGAUUAAUCAGCAUCUGAUUGUUUUCAAUUAGU---------------------GCCCGCGGCUGCGGCAGCUGUCAGCCGUCAAAUCCAAUAUAUGUAAAUUC-CCCC ........((((((((.....(((.(((((((....))))))))---------------------))..((((((((((...))).)))))))..))))))))............-.... ( -30.60) >DroSim_CAF1 16116 94 + 1 CUAUCAGCAUUGGAUUAAUCAGCAUCUGAUUGUUUUCAAUUAGU---------------------GCCCGCGGCUGCGGCAGCUGUCGGCCGUCAAAUCCAAUAU----AAAUCC-CCCC ........((((((((.....(((.(((((((....))))))))---------------------))..((((((((((...))).)))))))..))))))))..----......-.... ( -29.90) >DroEre_CAF1 16246 93 + 1 CUAUCAGCAUUGGAUUAAUCAGCAUCUGAUUGUUUUCAAUUAGU---------------------GCCU-CGACUCCGGCAGCUGUCAGCCGUCAAAUCCAAUAU----AAAUCC-CCCC ........((((((((.....(((.(((((((....))))))))---------------------))..-.(((...(((........)))))).))))))))..----......-.... ( -23.50) >DroYak_CAF1 17048 93 + 1 CUAUCAGCAUUGGAUUAAUCAGCAUCUGAUUGUUUUCAAUUAGU---------------------GCCC-AAACUACGGCAGCUGUCAGCCGUCAAAUCCAAUAU----AAAUCC-CCCC ........((((((((.....(((.(((((((....))))))))---------------------))..-.....(((((........)))))..))))))))..----......-.... ( -24.50) >DroAna_CAF1 448 116 + 1 UUAUCAGCAUCGGGCUAAUCAGCAUCUGAUUAUUUUCAAUUAGUUGGCCAGUUCGGGCUUACCCUGCCUAUGCAUUUAGCAGCUGUCAGUCCACU-AUCCAAUAUA--UAAAUCC-CCCC ...........(((.(((((((...)))))))..........(((((..(((..(((((.((.((((.((......))))))..)).))))))))-..)))))...--......)-)).. ( -26.10) >consensus CUAUCAGCAUUGGAUUAAUCAGCAUCUGAUUGUUUUCAAUUAGU_____________________GCCCGCGGCUGCGGCAGCUGUCAGCCGUCAAAUCCAAUAU____AAAUCC_CCCC ........((((((((...((((..(((((((....)))))))......................(((.........))).))))..........))))))))................. (-16.01 = -16.87 + 0.86)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:54 2006