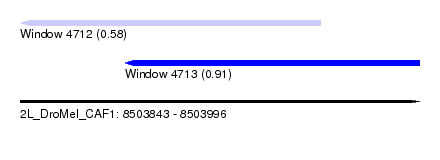
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,503,843 – 8,503,996 |
| Length | 153 |
| Max. P | 0.908374 |

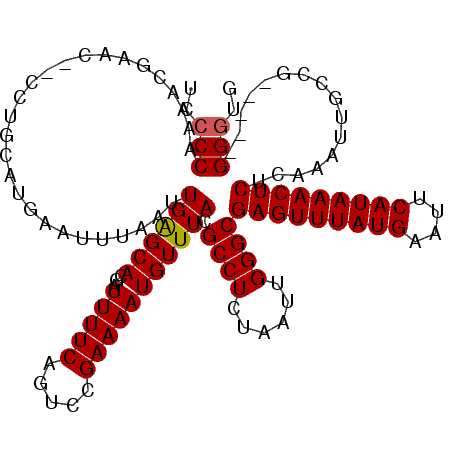
| Location | 8,503,843 – 8,503,958 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.56 |
| Mean single sequence MFE | -31.75 |
| Consensus MFE | -23.68 |
| Energy contribution | -24.24 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582659 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8503843 115 - 22407834 AUUUUCAGUCCGAAAAUGUUUACGCCUCUAAUUGGGCGAGUUUAUGAAUUCAUAAACUCUCAAAUUGCCG-----GGGUGUUAAAAGUCAAGUGAAGUGGGCUUUAUGUAGUGCGUAGCA ((((((.....))))))(((.((((..(((...(((((((((((((....)))))))))(((..(..(..-----((.(......).))..)..)..)))))))....))).))))))). ( -30.70) >DroSec_CAF1 13160 115 - 1 AUUUUCAGUCCGAAAAUGUUUACGCCUCUAAUUGGGCGAGUUUAUGAAUUCAUAAACUCUCAAAUUGCCG-----GGGUGUUAAAAGUCAAGUGAAGGGGGCUUUAUGUAGUGCGUAACA ......(((((..........((((((((((((..(.(((((((((....))))))))).).)))))..)-----))))))......((....))...)))))(((((.....))))).. ( -29.00) >DroSim_CAF1 12763 115 - 1 AUUUUCAGUCCGAAAAUGUUUACGCCUCUAAUUGGGCGAGUUUAUGAAUUCAUAAACUCUCAAAUUGCCG-----GGGUGUUAAAAGUCAAGUGAAGGGGGCUUUAUGUACUGCGAACUA ((((((.....))))))((((((((((((((((..(.(((((((((....))))))))).).)))))..)-----))))))....(((...((((((....))))))..)))..)))).. ( -30.20) >DroEre_CAF1 12757 114 - 1 AUUUUCAGUCCGAAAAUGUUUACGCCUCUAAUUGGGCGAGUUUAUGAAUUCAUAAACUCGCAAAUACCAG-----GGGUGUUAAAAGUCAAGUGAAG-GGGCUUUAUGUAGUGCGUAACA ((((((.....))))))......((((((..(((((((((((((((....))))))))))).((((((..-----.)))))).....))))....))-)))).(((((.....))))).. ( -35.70) >DroYak_CAF1 13514 119 - 1 AUUUUCAGUCCGAAAAUGUUUACGCCUCUAAUUGGGCGAGUUUAUGAAUUCAUAAACUCUCAAAUACCCAGGCCUGGAUGUUAAAAGUCAAACGAGG-GGGCUUUAUGUAGUGCGUAACA ((((((.....))))))..((((((..(((...(((((((((((((....))))))))).......(((.......(((.......))).......)-))))))....))).)))))).. ( -33.14) >consensus AUUUUCAGUCCGAAAAUGUUUACGCCUCUAAUUGGGCGAGUUUAUGAAUUCAUAAACUCUCAAAUUGCCG_____GGGUGUUAAAAGUCAAGUGAAG_GGGCUUUAUGUAGUGCGUAACA ((((((.....))))))..((((((..(((....((((((((((((....)))))))))........((......)).........)))..((((((....)))))).))).)))))).. (-23.68 = -24.24 + 0.56)

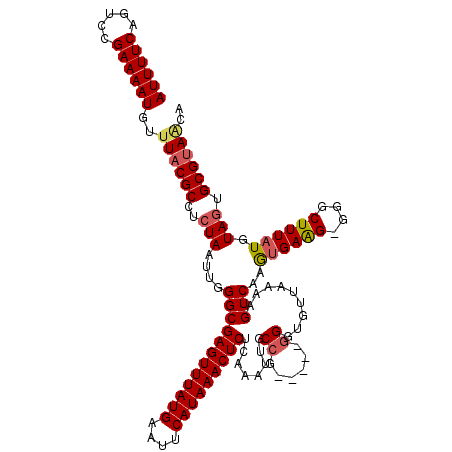
| Location | 8,503,883 – 8,503,996 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -31.24 |
| Consensus MFE | -23.62 |
| Energy contribution | -23.66 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8503883 113 - 22407834 UCCCCAAAACGAAC--CCUGCAUGAAUUUAAUUGAGCACGAUUUUCAGUCCGAAAAUGUUUACGCCUCUAAUUGGGCGAGUUUAUGAAUUCAUAAACUCUCAAAUUGCCG-----GGGUG ............((--(((((((((.......((((((...(((((.....))))))))))).((((......))))(((((((((....))))))))))))...)).))-----)))). ( -31.10) >DroSec_CAF1 13200 113 - 1 UCCCCAAAACGAAC--CCUGCAUGAAUUUAAUUGAGCACGAUUUUCAGUCCGAAAAUGUUUACGCCUCUAAUUGGGCGAGUUUAUGAAUUCAUAAACUCUCAAAUUGCCG-----GGGUG ............((--(((((((((.......((((((...(((((.....))))))))))).((((......))))(((((((((....))))))))))))...)).))-----)))). ( -31.10) >DroSim_CAF1 12803 113 - 1 UCCCCAAAAUGAAC--CCUGCAUGAAUUUAAUUGAGCACGAUUUUCAGUCCGAAAAUGUUUACGCCUCUAAUUGGGCGAGUUUAUGAAUUCAUAAACUCUCAAAUUGCCG-----GGGUG ............((--(((((((((.......((((((...(((((.....))))))))))).((((......))))(((((((((....))))))))))))...)).))-----)))). ( -31.10) >DroEre_CAF1 12796 113 - 1 --CCCCAAACGACCCUGCUGCAUGUAUUUAAUUGGGCACGAUUUUCAGUCCGAAAAUGUUUACGCCUCUAAUUGGGCGAGUUUAUGAAUUCAUAAACUCGCAAAUACCAG-----GGGUG --..........(((((......((((((....((((((.((((((.....))))))))....))))........(((((((((((....))))))))))))))))))))-----))... ( -34.60) >DroYak_CAF1 13553 117 - 1 --CCCCAAACGACU-GCCUGCAUGAAUUUAAUUGAGCACGAUUUUCAGUCCGAAAAUGUUUACGCCUCUAAUUGGGCGAGUUUAUGAAUUCAUAAACUCUCAAAUACCCAGGCCUGGAUG --............-(((((............((((((...(((((.....))))))))))).((((......))))(((((((((....))))))))).........)))))....... ( -28.30) >consensus UCCCCAAAACGAAC__CCUGCAUGAAUUUAAUUGAGCACGAUUUUCAGUCCGAAAAUGUUUACGCCUCUAAUUGGGCGAGUUUAUGAAUUCAUAAACUCUCAAAUUGCCG_____GGGUG ..(((...........................((((((...(((((.....))))))))))).((((......))))(((((((((....)))))))))................))).. (-23.62 = -23.66 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:50 2006