
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,503,457 – 8,503,616 |
| Length | 159 |
| Max. P | 0.962373 |

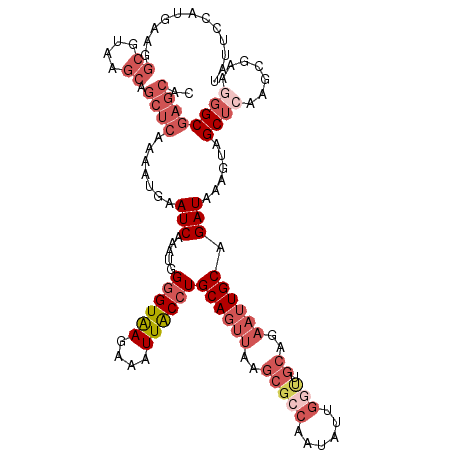
| Location | 8,503,457 – 8,503,577 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.50 |
| Mean single sequence MFE | -34.14 |
| Consensus MFE | -18.22 |
| Energy contribution | -22.38 |
| Covariance contribution | 4.16 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8503457 120 + 22407834 CAUUCCAUGACGGCGUAAGCAGCUCAAAAUGAAUCAAAUGGGGUAAGAAAUUACCUGCAGUUAAGCGCCAAUAUUGGUUGCAGAAUUGCAGAUAAAGUAGCUCAAGCGAAAGGCGAGCAC ((((...(((..((....))...))).)))).(((.....((((((....))))))((((((..((((((....))).)))..)))))).)))......((((...(....)..)))).. ( -34.70) >DroSec_CAF1 12841 120 + 1 UAUUUCAUGAAGGCGUAAGCAGCUCAAAAUGAAUCAAAUGGGGUAAGGAAUUACCUGCAGUUAAGCGCCCAUAUUGGUUGCAGAAUUGCAGAUAAAGUAGCUCAAGCGAUGGGCGAGCAU ............((....)).((((.......(((.....((((((....))))))((((((..(((.((.....)).)))..)))))).)))......(((((.....))))))))).. ( -36.10) >DroSim_CAF1 12373 120 + 1 UAUUCCAUGGCGGCGUAAGCAGCUCAAAAUGAAUCAAAUGGGGUAAGGAAUUACCUGCAGUUAAGCGGCAAUAUUGGUUGCAGAAUUGCAGAUAAAGUAGCUCAAGCGAUGGGCGAGCAC ............((....)).((((.......(((.....((((((....))))))((((((..(((((.......)))))..)))))).)))......(((((.....))))))))).. ( -36.20) >DroEre_CAF1 12449 115 + 1 UCUUCCAUCAAGGCGUAAGCAGCGCAGAAUUAAUCAAAUGG-GUAAGAAAUUGCCUGCAGUUGAGCGCCAAUAUUGGCUGCAGAAUUGCAGAUAGAGUAGCGCAACCGCAGGGCAG---- ...(((......((....)).((((....((.(((....((-((((....))))))((((((..((((((....)))).))..)))))).))).))...)))).......)))...---- ( -37.10) >DroYak_CAF1 13143 99 + 1 UCUUCCAUU---------CCAGCUCAGAAUGAAUCAAAUGGGGUGAGAAAUUACCUGCAGUUAAGCGCCAAUAU------------UGCAGAUAGAGUAGCUCAACCGAAAGGCGAGCAC .........---------...((((...............((((((....))))))(((((...........))------------))).....)))).((((..((....)).)))).. ( -26.60) >consensus UAUUCCAUGAAGGCGUAAGCAGCUCAAAAUGAAUCAAAUGGGGUAAGAAAUUACCUGCAGUUAAGCGCCAAUAUUGGUUGCAGAAUUGCAGAUAAAGUAGCUCAAGCGAAGGGCGAGCAC .....................((((.......(((.....((((((....))))))((((((..(((((......))).))..)))))).)))......((((.......)))))))).. (-18.22 = -22.38 + 4.16)


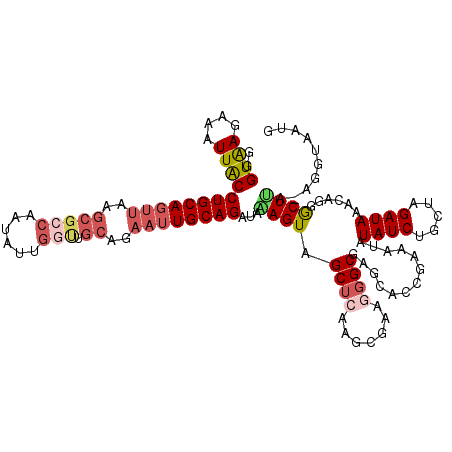
| Location | 8,503,497 – 8,503,616 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.70 |
| Mean single sequence MFE | -32.16 |
| Consensus MFE | -16.80 |
| Energy contribution | -19.72 |
| Covariance contribution | 2.92 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8503497 119 + 22407834 GGGUAAGAAAUUACCUGCAGUUAAGCGCCAAUAUUGGUUGCAGAAUUGCAGAUAAAGUAGCUCAAGCGAAAGGCGAGCACCGAAAUAUAUCUGCUAGAUAAACAGGGCUCA-AGGUACUG ((((((....))))))((((((..((((((....))).)))..))))))......((((.((...((.....))((((.((......((((.....))))....)))))).-)).)))). ( -36.50) >DroSec_CAF1 12881 119 + 1 GGGUAAGGAAUUACCUGCAGUUAAGCGCCCAUAUUGGUUGCAGAAUUGCAGAUAAAGUAGCUCAAGCGAUGGGCGAGCAUCGAAAUAUAUCUGCUAGAUAAACAUGGCUUA-AGUUAAAG ((((((....))))))((((((..(((.((.....)).)))..))))))........(((((.((((.(((..(.((((..((......)))))).).....))).)))).-)))))... ( -32.70) >DroSim_CAF1 12413 119 + 1 GGGUAAGGAAUUACCUGCAGUUAAGCGGCAAUAUUGGUUGCAGAAUUGCAGAUAAAGUAGCUCAAGCGAUGGGCGAGCACCGAAAUAUAUCUGCUAGAUAAACAGGGCUUA-AGUUAAAG ((((((....))))))((((((..(((((.......)))))..))))))..........(((((.....)))))((((.((......((((.....))))....)))))).-........ ( -31.50) >DroEre_CAF1 12489 114 + 1 G-GUAAGAAAUUGCCUGCAGUUGAGCGCCAAUAUUGGCUGCAGAAUUGCAGAUAGAGUAGCGCAACCGCAGGGCAG-----GAAAUGUAUCCGUUAGAUAAAUAGGGCUUAAAAGUAUUG (-((((....)))))(((..(((((((((........((((......))))........(((....)))..))).(-----((......)))..............))))))..)))... ( -31.50) >DroYak_CAF1 13174 108 + 1 GGGUGAGAAAUUACCUGCAGUUAAGCGCCAAUAU------------UGCAGAUAGAGUAGCUCAACCGAAAGGCGAGCACCGAAAUGUAUCUCUUAGAUAAAUAGUACUCACAGGUAUUG ((((((....))))))(((((...........))------------))).....(((((((((..((....)).)))).........((((.....)))).....))))).......... ( -28.60) >consensus GGGUAAGAAAUUACCUGCAGUUAAGCGCCAAUAUUGGUUGCAGAAUUGCAGAUAAAGUAGCUCAAGCGAAGGGCGAGCACCGAAAUAUAUCUGCUAGAUAAACAGGGCUUA_AGGUAAUG ..((((....))))((((((((..(((((......))).))..))))))))...((((.((((.......)))).............((((.....))))......)))).......... (-16.80 = -19.72 + 2.92)

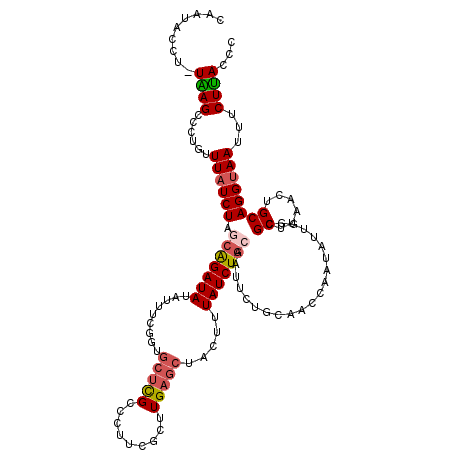
| Location | 8,503,497 – 8,503,616 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.70 |
| Mean single sequence MFE | -30.04 |
| Consensus MFE | -12.82 |
| Energy contribution | -13.50 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8503497 119 - 22407834 CAGUACCU-UGAGCCCUGUUUAUCUAGCAGAUAUAUUUCGGUGCUCGCCUUUCGCUUGAGCUACUUUAUCUGCAAUUCUGCAACCAAUAUUGGCGCUUAACUGCAGGUAAUUUCUUACCC .((((.((-..(((.(((((.....))))).........(((....)))....)))..)).))))(((((((((.((..((..(((....))).))..)).))))))))).......... ( -32.80) >DroSec_CAF1 12881 119 - 1 CUUUAACU-UAAGCCAUGUUUAUCUAGCAGAUAUAUUUCGAUGCUCGCCCAUCGCUUGAGCUACUUUAUCUGCAAUUCUGCAACCAAUAUGGGCGCUUAACUGCAGGUAAUUCCUUACCC ......((-(((((......((((.....))))......((((......))))))))))).....(((((((((.((..((..((.....))..))..)).))))))))).......... ( -28.50) >DroSim_CAF1 12413 119 - 1 CUUUAACU-UAAGCCCUGUUUAUCUAGCAGAUAUAUUUCGGUGCUCGCCCAUCGCUUGAGCUACUUUAUCUGCAAUUCUGCAACCAAUAUUGCCGCUUAACUGCAGGUAAUUCCUUACCC ......((-(((((.(((((.....))))).........(((....)))....))))))).....(((((((((.((..((.............))..)).))))))))).......... ( -26.12) >DroEre_CAF1 12489 114 - 1 CAAUACUUUUAAGCCCUAUUUAUCUAACGGAUACAUUUC-----CUGCCCUGCGGUUGCGCUACUCUAUCUGCAAUUCUGCAGCCAAUAUUGGCGCUCAACUGCAGGCAAUUUCUUAC-C .........((((...............(((......))-----)((((.((((((((.((.........((((....))))((((....)))))).))))))))))))....)))).-. ( -33.60) >DroYak_CAF1 13174 108 - 1 CAAUACCUGUGAGUACUAUUUAUCUAAGAGAUACAUUUCGGUGCUCGCCUUUCGGUUGAGCUACUCUAUCUGCA------------AUAUUGGCGCUUAACUGCAGGUAAUUUCUCACCC ...((((((((((((((((.((((.....)))).))...))))))).......((((((((..(..(((.....------------)))..)..)))))))))))))))........... ( -29.20) >consensus CAAUACCU_UAAGCCCUGUUUAUCUAGCAGAUAUAUUUCGGUGCUCGCCCUUCGCUUGAGCUACUUUAUCUGCAAUUCUGCAACCAAUAUUGGCGCUUAACUGCAGGUAAUUUCUUACCC .........((((......((((((.(((((((.........(((((.........))))).....))))))).....................((......))))))))...))))... (-12.82 = -13.50 + 0.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:48 2006