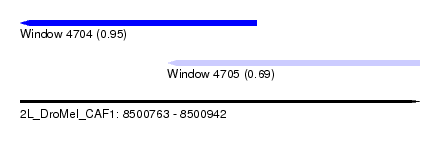
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,500,763 – 8,500,942 |
| Length | 179 |
| Max. P | 0.953521 |

| Location | 8,500,763 – 8,500,869 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.58 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -19.68 |
| Energy contribution | -19.96 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8500763 106 - 22407834 AACUCCCAGGAGGAUGUUAUCCUGUCGGUUGUUGUGGGCAAUGCUUUUAGCCUCCGCAAGUUAAAUGCUCCAACCU--------------CCAACCCCCUUUUGGAUAUCCUCCUCACCU .......(((((((((((.....(..((((((((.(((((..((((...((....))))))....)))))))))..--------------..))))..).....)))))))))))..... ( -31.50) >DroSec_CAF1 10206 119 - 1 AACUCCCAGGAGGAUGUUAUCCUGUCGGUUGUCGUGGGCAAUGCUUUUAGC-UCCGCAAGUUAAAUGCUCCAAACCCCUUUCCCCCAACCCCAAUCCCCUUUUGGAUUUCCUCCUCACCU .......(((((((............(((((..(.(((....((.((((((-(.....))))))).)).......)))....)..)))))..(((((......))))))))))))..... ( -29.30) >DroSim_CAF1 9415 119 - 1 AACUCCCAGGAGGAUGUUAUCCUGUCGGUUGUUGUGGGCAAUGCUUUUAGCCUCCGCAAGUUAAAUGCUCCAACCCCCUUUCCCCC-UCCCCAAUCCCCUUUUGGAUUUCCUCCUCACCU .......(((((((....((((....((..((((.(((....((.((((((........)))))).))..................-.)))))))..))....)))).)))))))..... ( -28.80) >DroEre_CAF1 9833 86 - 1 AACUCCCAGGAGGAUGUUAUCCUGUCGGUUGUUGUGGGCAAUGCUUUUAGCCUCCGCAAGUUAAAUGCUCCGACAA----GC------------------------------CCUCACCU ......(((((........)))))..((((((((.(((((..((((...((....))))))....))))))))).)----))------------------------------)....... ( -22.40) >DroYak_CAF1 10404 99 - 1 AACUCCCAGGAGGACGUUAUCCUGUCGGUUGUUGUGGGCAAUGCUUUUAGCCUCCGCAAGUUAAAUGCUCCAACCU----UCCCCC-----------------GGAUUUGCUCCUCACCU .........(((((.((.((((.(..((..((((.(((((..((((...((....))))))....)))))))))..----.))..)-----------------))))..))))))).... ( -27.40) >consensus AACUCCCAGGAGGAUGUUAUCCUGUCGGUUGUUGUGGGCAAUGCUUUUAGCCUCCGCAAGUUAAAUGCUCCAACCC____UCCCCC____CCAA_CCCCUUUUGGAUUUCCUCCUCACCU ((((..(((((........)))))..))))((((.(((((..((.....))...(....).....))))))))).............................................. (-19.68 = -19.96 + 0.28)

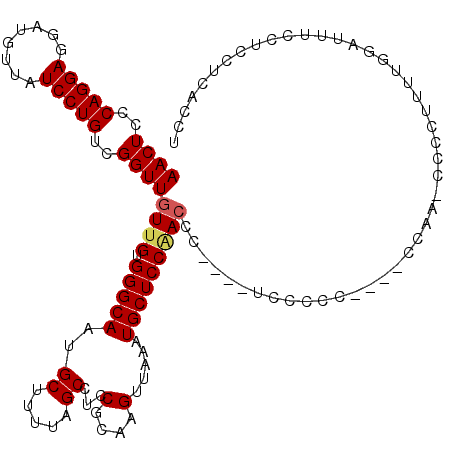

| Location | 8,500,829 – 8,500,942 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.20 |
| Mean single sequence MFE | -36.44 |
| Consensus MFE | -30.72 |
| Energy contribution | -31.36 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8500829 113 - 22407834 UCCAAA------UCCACGUUCUCAUCCACGUCCUGAAAGUCUCGGUCUCGAAAGAAACCCAAGGCGGCGCUCGCGUG-GCAACUCCCAGGAGGAUGUUAUCCUGUCGGUUGUUGUGGGCA ......------((((((.......((((((..((...((((.(((.((....)).)))..))))..))...)))))-)(((((..(((((........)))))..))))).)))))).. ( -38.60) >DroSec_CAF1 10285 107 - 1 ------------UCCACGUCCUCAUCCACGUCCUGAAAGUCUCGUUCUCGAAAGAAACCCAAGGCGCCGCUCGCGUG-GCAACUCCCAGGAGGAUGUUAUCCUGUCGGUUGUCGUGGGCA ------------((((((.(..(((((...(((((...((((.(((.((....)))))...))))(((((....)))-))......))))))))))....((....))..).)))))).. ( -36.00) >DroSim_CAF1 9494 108 - 1 ------------NNNNNNNNNNNNNNNNNNNNCUGAAAGUCUCGGUCUCGAAAGAAACCCAAGGCGGCGCUCGCGUGGGCAACUCCCAGGAGGAUGUUAUCCUGUCGGUUGUUGUGGGCA ------------..........................((((.(((.((....)).)))..))))...(((((((...((((((..(((((........)))))..))))))))))))). ( -35.20) >DroEre_CAF1 9879 95 - 1 UCCACGUC------------------------CUGAAAGGCUCGGUCUCGCAAGAAACCCAAGGCGGCGCUCGCGUG-GCAACUCCCAGGAGGAUGUUAUCCUGUCGGUUGUUGUGGGCA ....((((------------------------((....))...(((.((....)).)))...))))..(((((((..-((((((..(((((........)))))..))))))))))))). ( -36.30) >DroYak_CAF1 10463 119 - 1 UCCACGUCCUCAUCCUCAUCCUCUUCCAUGACCUGAAAGUCUCUGUGCCGACAGAAACCCAAGGCGGCGCUCGCGUG-GCAACUCCCAGGAGGACGUUAUCCUGUCGGUUGUUGUGGGCA ....((((.(((...((((........))))..)))..((.(((((....))))).))....))))..(((((((..-((((((..(((((........)))))..))))))))))))). ( -36.10) >consensus UCCA________UCC_C_U_CUC_UCCA_G_CCUGAAAGUCUCGGUCUCGAAAGAAACCCAAGGCGGCGCUCGCGUG_GCAACUCCCAGGAGGAUGUUAUCCUGUCGGUUGUUGUGGGCA ......................................((((.(((.((....)).)))..))))...(((((((...((((((..(((((........)))))..))))))))))))). (-30.72 = -31.36 + 0.64)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:42 2006